Human FEN1-PCNA Complex
Maria Narvaez '14 and Hailey Schneider '14
Contents:
I. Introduction
Flap endonuclease-1 (FEN1) is an enzyme involved in the
maintenance of genomic stability and replication. During replication
of lagging strand DNA, FEN1 aids in removing DNA primers during
Okazaki fragment maturation. FEN1 also plays an important role in long
patch base excision repair. Studies have shown that the interaction of
FEN1 with proliferating cell nuclear antigen (PCNA; the DNA sliding
clamp) stimulates the activity of FEN1, leading to a 10- to 50-fold
increase in its nuclease activity.
Mutations in human FEN1 impairing its endonuclease activity
have been linked to cancer development, which makes sense
considering its involvement in DNA replication and repair
processes.
II. General Structure
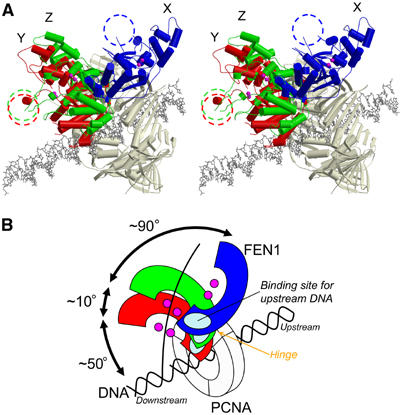
The human FEN1-PCNA complex is composed of 3 FEN1 molecules,
bound to 1 PCNA trimer. The three PCNA subunits tightly
associate to form a closed ring, with each subunit exhibiting
symmetry.
FEN1 and PCNA interact through beta-beta
previous button is new!!!!!!!!!!!!!!!
and hydrophobic interactions and these interactions keep the enzyme in
an inactive locked-down orientation. It is possible that these
interactions play a role in making rapid DNA-tracking possible by
preserving the central hole of PCNA as it slides along the DNA.
Human FEN1 has two domains, the
nucleus core domain and the C-terminal
tail domain.
The central groove of the core domain is formed by a group
of beta sheets with two helical
regions on both sides.
III. Activation
The human FEN1 active site, located at the central cleft, is
formed by two clusters of conserved acidic residues that bind two metal ions.
The first of these metal ion-binding sites is composed of Asp34,
Asp86, Glu158,
and Glu160. It is involved in the
catalysis of nucleophilic attack of the phosphodiester bond of DNA.
The second of the metal ion-binding sites is composed of Asp179, Asp181;
and Asp233. This site is thought to be
involved in DNA binding.
The active site also involves the interaction of a conserved Tyr234
residue with Glu158, via a hydrogen
bond, and with Asp181, via a water
mediated contact.
The core domain of FEN1 is linked to the PCNA-binding tail by a
short linker that acts as a hinge directing the FEN1 core domain
towards the DNA substrate.
IV. DNA Binding
FEN1 interacts with the template DNA strand at two physically
seperate regions, the H2TH:K+ ion and four basic residues: Arg239,
Lys244, Arg245, and Lys267, which makes up the largest FEN1:DNA
interface.
The clamp region,
which is believed to thread along the single strand DNA,
contains many charged and hydrophobic residues. In addition to these
phosphate interactions, the side chains of Glu-181
and Arg-185, both emanating from the recognition
helix
directly contact the bases within the major groove of the DNA. Because
of the way that the protein binds to the DNA, a kink of ~40 degrees
occurs between nucleotide base pairs six
and seven on each side of the dyad
axis, 5'-TG-3'
This sequence has been shown to favor DNA flexibility and bending in
other systems as well. Because of this kink, an additional five
ionic interactions and four hydrogen bonds are able to occur between
the protein and the DNA strand. Examples of these new interactions
occur between Lys-26, Lys-166, His-199
and the DNA sugar-phosphate backbone
The DNA bend is integral to the mechanism of transcription activation.
Not only does it place CAP in the proper orientation for interaction
with RNA polymerase, but wrapping the DNA around the protein may
result in direct contacts between upstream DNA and RNA
polymerase.
V. Complex Disruption
A double mutation involving Pro253 (and Lys254) in yeast has
been shown to affect the stimulation of FEN1-cite this?, with Pro253
thought to be involved in coordinating two oxygen atoms in the
backbone of Ala252 and Pro253 toward betaA of FEN1.
andTranscription activation by CAP requires more than merely
the binding of cAMP and binding and bending of DNA. CAP contains an
"activating region" that has been proposed to participate in direct
protein-protein interactions with RNA polymerase and/or other basal
transcription factors. Specifically, amino acids 156,
158, 159,
and 162
have been proposed to be critical for transcription activation by CAP.
These amino acids are part of a surface loop composed of residues
152-166
Researchers have concluded that the third and final step in
transcription activation is this direct protein-protein contact
between amino acids 156-162 of CAP, and RNA polymerase.
VI. References
Tsutakawa, Susan E., Scott Classen, Brian R.
Chapados, Andrew S. Arvai, L. David Finger, Grant Guenther,
Christopher G. Tomlinson, Peter Thompson, Altaf H. Sarker, Binghui
Shen, Priscilla K. Cooper, Jane A. Grasby, and John A. Tainer. 2011.
Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a
Unified Understanding of the FEN1 Superfamily. Cell
145:198-211.
Sakurai, Shigeru, Ken Kitano, Hiroto
Yamaguchi, Keisuke Fukuda, Makiyo Uchida, Eiko Ohtsuka, Hiroshi
Morioka, and Toshio Hakoshima. 2005. Structral Basis for
Recruitment of Human Flap Endonuclease 1 to PCNA. The EMBO
Journal 24.4: 683-93.
Vaney, Marie Christine, Gary L. Gilliland,
James G. Harman, Alan Peterkofsky, and Irene T. Weber. 1989.
Crystal Structure of a cAMP-Independent Form of Catabolite Gene
Activator Protein with Adenosine Substituted in One of Two
cAMP-Binding Sites. Biochemistry 28:4568-4574.
Weber, Irene T., Gary L. Gilliland, James G.
Harman, and Alan Peterkofsky. 1987. Crystal Structure of a Cyclic
AMP-independent Mutant of Catabolite Activator Protein. The
Journal of Biological Chemistry 262:5630-5636.
Zhou, Yuhong, Ziaoping Zhang, and Richard H.
Ebright. 1993. Identification of the activating region of
catabolite gene activator protein (CAP): Isolation and
characterization of mutants of CAP specifically defective in
transcription activation. Proceedings of the National Academy
of Sciences of the United States of America 90:6081-6085.
Back to Top