CueR: A
Copper Efflux Regulator in Escherichia coli
Carolina Andrade '19 and Bryce Nicholls '18
Contents:
Model View:
Color Scheme:
I. Introduction
The MerR protein family is a group of transcription factors
characterized by N-terminal helix-turn-helix DNA binding domains and
C-terminal effector regions. Found widely in prokaryotes, MerR
proteins are sensitive to a variety of environmental stimuli
including antibiotics, heavy metals, and oxidative stress, but only
a small subgroup of MerRs are sensitive to metal ions [1-2].
Members of this subgroup, which includes MerR itself, are called
metalloregulators, and they modulate transcription by distorting DNA
conformation in response to metal-binding [3].
Copper efflux regulator (CueR) is a Cu+- and Ag+-sensing
metalloregulator that controls the expression of two genes involved
in metal homeostasis: CopA, which encodes a copper/silver
ATPase, and CueO, which encodes a copper oxidase [2].
Like most other metalloregulators, CueR acts on promoter DNA that
exceeds the optimal length (~17bp) for recognition by sigma70, a
subunit of RNA polymerase (RNAP) [1,2].
In its repressor form, CueR bends DNA at acute angles,
stereochemically preventing RNAP from binding to both the -10 and
-35 regions of the promoter [1]. However, because CueR readily binds
copper (KD= 2x10-21M), CueR is most often found in the activator
form [1]. In fact, its metal
affinity is so high that Philips et al. (2015) had to mutate the
metal-binding residues in the AgI-CueR complex in order to create
and study a constitutive repressor.
Once Cu or Ag binds to CueR, the protein
enters the activator state, which introduces torsional stress on the
DNA that brings the -10 and -35 regions into better contact with
RNAP and thus optimizes RNAP-promoter binding. Through this dynamic
command of promoter DNA conformation, CueR and its metal ligands are
able to act as an ‘on/off’ switch for DNA transcription.
II.General Structure
CueR is a
made up of ten
five in each monomer. Dimerization occurs on the C’ terminal end
through interactions between each monomer’s fifth alpha helix, aptly
called the
(DH). Through a
the DH connects to a
(DBD) on the N-terminal of the
protein. The DBD is composed of four alpha helices (a1, a2, a3, and
a4) arranged in a winged helix-turn-helix motif [1,2].
The ‘wing’ portion of the winged helix-turn-helix motif is composed
of small
which also interact with promoter DNA [1,4].
In the activator state, the dimerization helix is followed by
a metal-binding loop (MBL) and
a sixth alpha helix called the two turn C-terminal alpha helix (CTH).
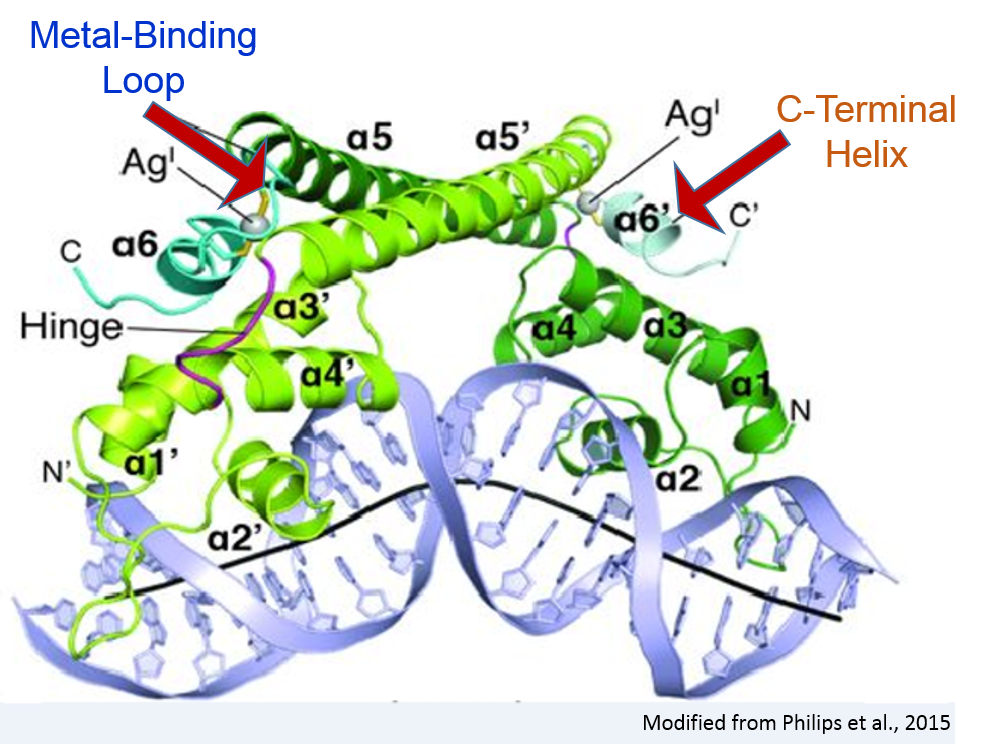
Both of these structures are disordered in the repressor.
III. DNA Binding
CueR recognizes DNA with
which inserts into the minor groove and hydrogen bonds with the
second nitrogen in the guanine at the 22nd position in one monomer
and G23 in the other monomer (henceforth denoted as prime “ ‘ ”), as
well as with an oxygen from T23/T24'.
is also involved in DNA recognition. It inserts into the major
groove and hydrogen bonds with the nitrogen and oxygen of G18/19'
and the nitrogen of G17/18'. In both forms of CueR, the aromatic
ring of
participates in van der Waals interactions. These van der Waals are
crucial for stacking with DNA bases,and despite existing in both the
activator and repressor forms, these van der Waals forces have a
greater role in stability in CueR's active state [1].
The conserved van der Waals are not an anomaly; many of the
CueR-DNA contacts between the DBD
and the copA promoter are conserved in both the activator
and repressor states. Notably, the
a structure that consists of three arginine residues (Arg-18,
Arg-31, and Arg-37) and the wing loop of the DBD,
interacts with the phosphate groups of the DNA backbone in both the
repressor and activator states. This "R Clamp" is thought to be
responsible for distorting promoter DNA into A-DNA conformation, a
topological change critical for transcriptional activation.
IV. Allostery and Regulation of DNA Topology
The greatest difference between the repressor and activator
is the conformation and spacing of the bound promoter DNA. The copA
promoter is ~19 bp long between the -10 and -35 regions. In the
repressor form, the seven central basepairs within the ~19 bp are in
conformation. Once a metal ligand binds, the central DNA bends by 36
degrees and enters an 'A'-DNA-like conformation called TA-DNA. The
the central basepairs (T12&T13’ and T13&G14’) also become
in response to the widening of the minor groove [1].
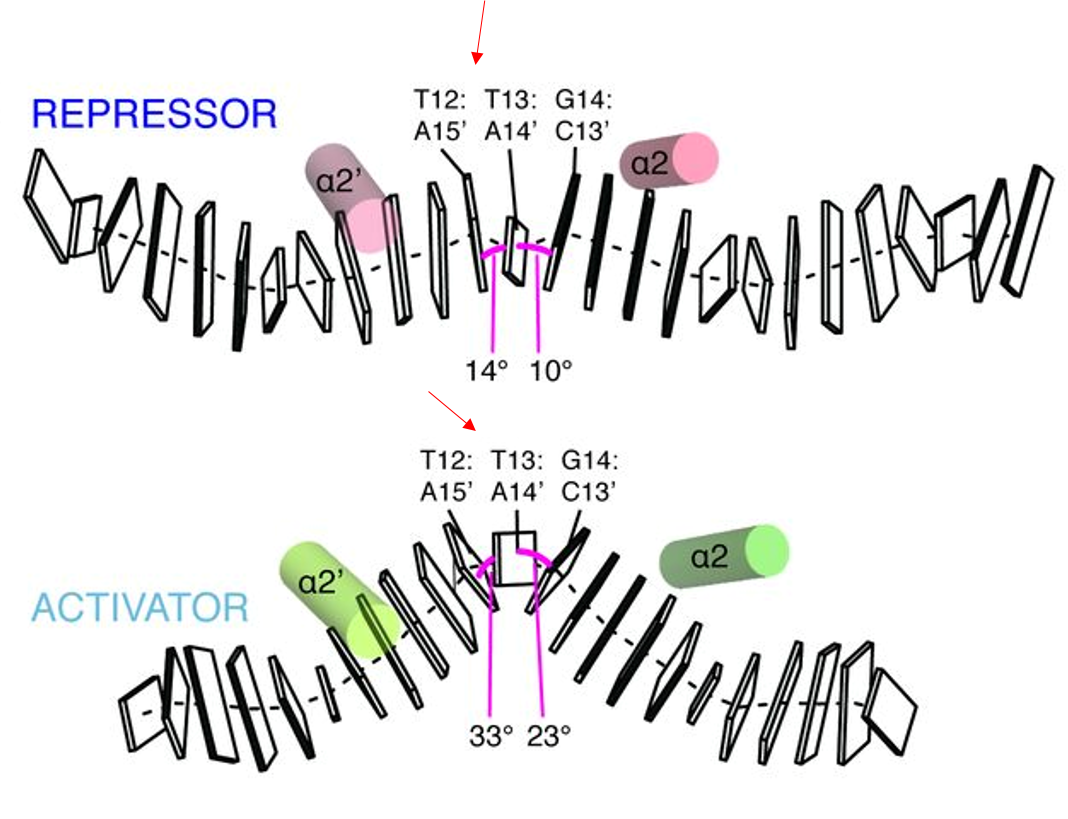
These conformational changes occur as a result of metal
binding. When a metal ligand binds to CueR, it assembles the
metal-binding loop (MBL) and
the C-terminal helix (CTH),
which were both disordered in the repressor form.
In doing so, Ala118’ of the MBL
hydrogen bonds with Arg 75 in the hinge loop, causing the arginine
residue to flip out and displace a water molecule [1].
In its new position, Arg75 acts as a link between the MBL
and the DBD, where Philips et
al. (2015) suggests it plays an important role in stability by
establishing hydrogen bond communication between the subunits.
Because transcription decreased in complexes with mutant Arg75 [1], the residue and the
communication it mediates are thought to be crucial to
transcriptional activation.
The CTH further
stabilizes the activator state by interacting with
.
Ile122, Ile123,
and Leu126
of the CTH will insert into
the cavity, causing Phe70)'s
aromatic ring to flip out towards a4. This displacement forces the
dimerization helices DHs
(a5 and a5’) to "scissor", locking the complex in a tighter
orientation and decreasing the distance between the DBDs
in the process [1]. Since the
DBDs are bound tightly to the
phosphate groups of the DNA backbone, pulling them together results
in the kinking and undertwisting of promoter DNA, decreasing the
distance by 6 angstroms.
CueR's ability to optimize promoter length in the active
form by retaining the protein-DNA contacts present in the repressor
confers an advantage. It allows E. coli to survive in
environments with elevated concentrations of copper, which would
otherwise have toxic effects, without expending energy [5].
V. References
1. Phillips, Canalizo-Hernande,Yidirim,
Schatz, Mondragon, O'Hallran. 2015. Allosteric transcription
regulation via changes in the overall topology of the core
promoter. Science. Vol 349:6250:877-881.
2. Brown, N.L., Stoyanov, J.V., Kidd, S.P.,
Hobman, J.L. 2003. The MerR family of transcriptional
regulators. FEMS Microbiology Reviews. Vol
27:2-3:145-163.
3. Chandra P. Joshi, Debashis Panda, Danya
J. Martell, Nesha May Andoy, Tai-Yen Chen, Ahmed Gaballa, John
D. Helmann, and Peng Chen 2012. Direct substitution and assisted
dissociation pathways for turning off transcription by a
MerR-family metalloregulator PNAS. 109 (38) 15121-15126
4. Brennan, R. G., (1993). The Winged-Helix
DNA-Binding Motif: Another Helix-Turn-Helix Takeoff. Cell.
Vol. 74, 773-776.
5. Grass, Gregor, and Christopher Rensing.
“Genes Involved in Copper Homeostasis in Escherichia Coli.” Journal
of Bacteriology. 183.6 (2001): 2145–2147.
Back to Top