E. coli Multiple
Antibiotic Resistance Regulator Protein
Erick Ditmars '18 and Kay Burrows '18
Contents:
I. Introduction
The Escherichia coli Multiple antibiotic resistance
regulator protein (MarR) is the hallmark member of the MarR family,
responsible for regulating genes involved in antibiotic resistance.
The MarR family of proteins is found in both prokaryotes and archaea
and have been shown to play a significant role in the development of
antibiotic resistant bacterial strains that pose a severe threat to
global healthcare. E. coli's MarR protein acts as a negative repressor
of the MarRAB system
When this system is activated, its primary active protein,
MarA, acts as a transcriptional factor, upregulating 60 resistance
genes. These genes typically encode for proteins involved in the
construction of multi-drug efflux pumps, which allowing the
organism to resist not only antibiotics, but also other molecules
such as oxidative agents or disinfectants.
In its native form, the MarR protein binds to the Mar
operator upstream of the marA gene. When bound, the protein acts
as a steric hinderance to RNA polymerase, blocking transcription.
MarR also has a strong affinity to aromatic acid molecules that,
when bound, inactivate the operator binding affinity of the MarR
protein. This lifts the steric block, allowing for the
transcription of MarA and the formation of the cell's antibiotic
resistance phenotype.
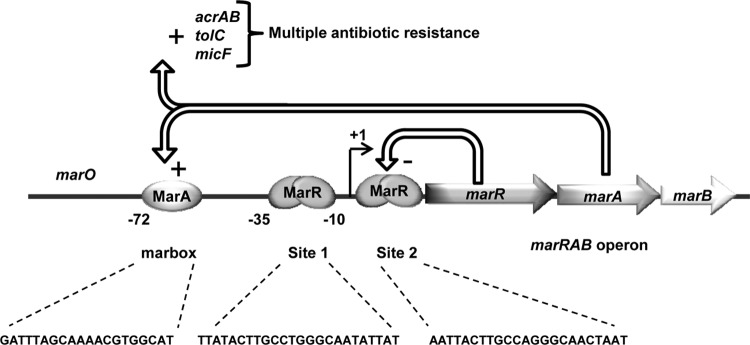
Figure 1. The MarRAB operon by Duval et al.
II. General Structure
MarR is a
comprised of two identical polypeptide chains with approximate overall
dimensions of 50 X 55 X 45 Angstroms. Each subunit
is made up of 6
and three beta pleated
. The two subunits that form the dimer are held together by a linkage
between the
domain and the
domain. In
the N-terminal and C-terminal
domains form a linker between the subunits of the
protein, stabilizing the complex. The dimer is
further stabilized in this region through hydrogen
bonding between
as well as interactions between
. Another important linker between the two subunits comes from the
interaction of
which attract each other, holding the complex together.
III. DNA binding
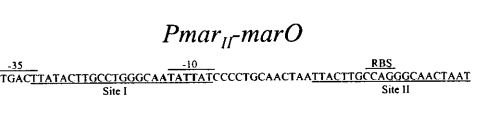
Figure 1. The MarR binding sites by Alekshun et
al.
While each of the precise
interactions have yet to be identified, many
studies have concluded that amino acids 61-121 of
MarR form a DNA binding domain using the
binding motif. The
motif binds to the phosphodiester backbone, stabalizing the
complex while the
motif binds within the major groove and is used for sequence
recognition of two sites on MarO. At this point in
time the exact method of protein-DNA interaction
in this protein is unknown, but a
mutation within the a4
helix eliminates the proteins DNA binding ability.
In addition, a "superrepressor" mutation of
, between the a4 and a5
helices; as been shown to increase binding by
upwards of 30 times. (G95 on one subunit could not
be shown due to errors in data collection using
the electron microscope). While these amino acids
are known to play a role in binding, the specific
binding motif is unknown. Currently, there are two
competeing theroies on how MarR binds with DNA.
The first model is predicated off of the
winged-helix binding of the E2F-DP transcriptional
factor heterodimer. In this model, MarR binds in
two half-faces, one per each subunit. These
half-faces bind on diffrent faces of the DNA
double helix. Both of these half-faces bind with
the major groove and play a role in sequence
recognition. The second binding motif is
reminiscent of DtxR binding. This second binding
motif is similar to that of E2F-DP except that the
half faces bind on the same face of the DNA helix.
IV. Salicylate binding
Sodium
salicylate, a model inhibitor for MarR, is
found to disrupt the protein’s activity in vitro
and in cells, and therefore can be used to
identify potential ligand binding domains in the
protein. Some studies hypothesize that
salicylate binds two sites on a single subunit
of MarR, SAL-A and SAL-B, located on either side
of the
In the proposed
, the interaction is comprised of hydrogen bonds with Thr 72
and Arg 86, with the salicylate ring positioned
over the hydrophobic chain of Pro 57. Similarly,
in the proposed
, the salicylate interacts through hydrogen bonds with Ala 70
and Arg 77 , with a hydrophobic Met 74 located
under the salicylate ring. Due to their
proximity to the DNA-binding helices, both sites
are hypothesized to cause a conformational
change in the DNA binding domains, disrupting
binding activity and rendering the MarR molecule
inactive.
However, a study by Duval et. al (2013) demonstrate
that mutations in these regions did not result in a
change in salicylate binding, suggesting these are
not the physiological binding sites, but rather
simply sites important for DNA binding. This study,
through mutagenesis, suggests that
were of greater importance to salicylate binding. These
newly identified amino acids create a hypothesized
ligand binding site between the DNA
binding domain and the N-terminal
and C-terminal
dimerization domains.
VI. References
Alekshun MN, Levy SB, Mealy
TR, Seaton BA, Head JF. The crystal structure of
MarR, a regulator of multiple antibiotic
resistance, at 2.3 A resolution. Nat Struct
Biol. 2001 Aug;8(8):710-4.
Duval V, McMurry LM, Foster
K, Head JF, Levy SB. A mutational analysis of
the multiple antibiotic resistance regulator
MarR reveals a ligand binding pocket at the
interface between the dimerization and
DNA-binding domains. J Bacteriol.
2013;195:3341–3351.253: 1001-1007.
Wilkinson, S. P. and Grove,
A. ( 2006 ). Ligand-responsive transcriptional
regulation by members of the MarR family of
winged helix proteins. Curr Issues Mol Biol
8, 51–62.
Martin R.G., Rosner J.L
(1995) Binding of purified multiple antibiotic
resistance repressor protein (MarR) to mar
operator sequences. Proc. Natl. Acad. Sci.
USA 92, 5456–5460.
Back to Top