Halobacterium salinarum Bacteriorhodopsin
Protein
Jessie Griffith '19 and Karina Kunka '19
Contents:
I. Introduction
Halobacterium salinarum is a halophilic archaeon found where sodium chloride concentrations reach 4.3 M, or seven times the concentration of seawater (Oren, 2006). Halobacterium salinarum is considered a polyextremophile, given its ability to also survive high levels of solar radiation and temperature stress such as is found in Qarhan Salt Lake (Xie et al., 2017).
Many species of halophilic archaea, including Halobacterium salinarum, produce photosensory proteins which aid in phototactic movement. A close homolog of these sensory rhodopsins is the photon-driven proton pump bacteriorhodopsin.
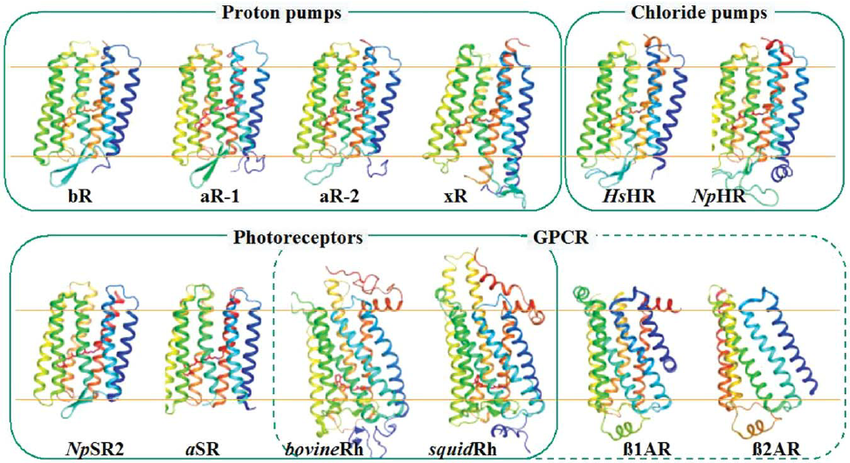 is abbreviated bR in the figure (pictured above). The sensory rhodopsin from anabaena, another unicellular organism, is abbreviated as aSR.
is abbreviated bR in the figure (pictured above). The sensory rhodopsin from anabaena, another unicellular organism, is abbreviated as aSR.
Bacteriorhodopsin pumps protons into the extracellular space in order to generate proton motive force (PMF). PMF is essential for both ATP production and mitigation of pH stress.
II. General Structure
Bacteriorhodopsin is a
and is the best-understood ion pump. It is the structural prototype of G protein-coupled receptors that contain transmembrane α-helices.
Each
in the trimer has seven membrane-spanning
The polypeptide is color-coded from N-terminus to C-terminus .
Helix color code: A B C D E F G
The loops between helices in the monomer also form a twisted anti-parallel β-sheet. This
is stabilized by six hydrogen bonds and faces the
The monomer is extensively stabilized by 31 hydrogen bonds. Of these 31 contacts,
can be ascribed to side-chain/main-chain and side-chain/side-chain hydrogen bonds between neighboring helices.
These hydrogen bonds contribute to the stability of the barrel-shaped biological unit. In addition, there are hydrogen bonds between non-neighboring helices, and these account for the remaining 12 contacts. Taken together, the stability from the hydrogen bonds allows each monomer to function as an independent proton pump, even before formation of quaternary structure. The units are held in the rigid purple membrane lattice.
III. Lipid Contacts
Eighteen tightly held lipid chains form a ring-shaped
about the co-crystallized protein. Nearly all of the contacts between the trimer and the other membrane components are mediated by these
The purple membrane contains five differnet types of lipids, all of which may make contacts with bacteriorhodppsin. For each monomer of bacteriorhodopsin, the purple membrane has proportionally five phosphatidyl glycerophosphates, two glycolipid sulfates, 0.5 phosphatidyl glycerol, 0.5 phosphatidyl glycerosulfate, as well as one molecule of
Squalene is the most hydrophobic membrane lipid, and is thought to modulate an electrostatic repulsion between the nearby charged Aspartic acid residues
of the cytoplasmic loop region and the membrane surface of the inner trimer space. This repulsion helps induce mobility in the α-helices to better accomodate isomerization of retinal and therefore increases the speed of the photocycle (Hendler et al., 2003). See " IV. Retinal Binding" below for more information.
IV. Retinal Binding
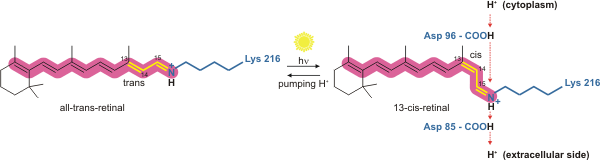
The retinal molecule, pictured above in both its all-trans and 13-cis form, consists of a polyene chain and a β-ionone ring. The final carbon shown (Carbon 15) is actually double bonded to the sidechain of
When the retinal molecule absorbs the energy of a photon given off by the sun, the C13=C14 double bond isomerizes from trans to cis.
Overall, the molecule interacts with a total of 12 amino acid side chains that collectively stabilize binding at the
The trimer contains three retinal binding pockets, allowing multiple retinal molecules to capture photons in the assembled protein.
Electrostatic interactions between retinal and the binding pocket are depicted.
V. Proton Translocation
As mentioned before, the 15th carbon of the retinal molecule forms a double bond with the Nitrogen in the sidechain of
This linkage is called a Schiff base. A Schiff base is an imine with 3 R groups where R3 must be an alkyl or an aryl group, and R1 and R2 may not be hydrogens.
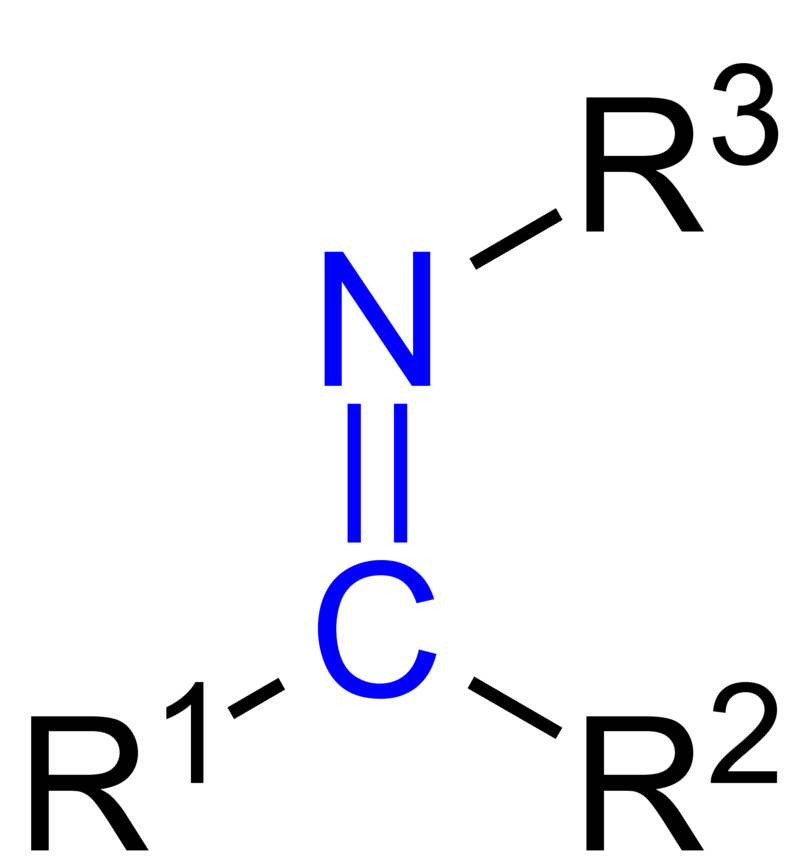
The Schiff base of the retinal molecule is mainly comprised of the covalent link between Lys216 and the chromophore. It is formed by the nucleophilic attack of the amine nitrogen of Lys216 on the carbonyl carbon of retinal. This linking of retinal to the binding pocket via Lys216 anchors retinal in place as it undergoes photoisomerization upon absorption of a photon.
There is an extensive
that stabilizes separated charges around the active site composed of the retinal Schiff base, Asp85, Asp212, and waters 401, 402, 406, and 407.
Photoisomerization results in release of a proton which is then preferentially accepted by Asp85. Another potential proton acceptor, Asp212, is stabilized in its anionic form by
with two sterically hindered Tyr residues, and thus is less likely to accept a proton.
Protonation of Asp85 releases a proton from a holding site into the extracellular space. The
is comprised of waters 403, 404, and 405, and glutamate 194 and 204. The holding site is shielded from the extracellular aqueous environment by Ile78 and Leu201.
Asp96 restores retinal to its original isomeric trans form by reprotonation, leading to release of a second proton to the extracellular space. Asp85 releases the proton it accepted from isomerized retinal into the holding site so the process can begin again.
This means that for each cycle of trans-cis-trans retinal , two protons are transported to the extracellular space.
VI. References
D. (2012, October 2). Bacteriorhodopsin retinal [Digital image]. Retrieved from https://upload.wikimedia.org/wikipedia/commons/5/57/Bacteriorhodopsin_retinal.png
J. (2010, March 28). Imino Group [Digital image]. Retrieved from https://upload.wikimedia.org/wikipedia/commons/thumb/7/72/Imino_Group_V.1.png/800px-Imino_Group_V.1.png
Luecke, H., Schobert, B., Richter, H., Cartailler, J., and Lanyi, J. (1999). Structure of bacteriorhodopsin at 1.55 A resolution. Journal of Molecular Biology, 291(4), 899-911. doi:10.1006/jmbi.1999.3027
Okada, T., and Kouyama, T. (1996, January). Structures of bacteriorhodopsin bR pdb id 1iw6 archaerhodopsin 1 and 2 aR1 [Digital image]. Retrieved from Structures-of-bacteriorhodopsin-bR-pdb-id-1iw6-archaerhodopsin-1-and-2-aR1
Oren, A. (2006). Life at High Salt Concentrations. The Prokaryotes, 263-282. doi:10.1007/0-387-30742-7_9
Xie, K., Deng, Y., Zhang, S., Zhang, W., Liu, J., Xie, Y., . . . Huang, H. (2017). Prokaryotic Community Distribution along an Ecological Gradient of Salinity in Surface and Subsurface Saline Soils. Scientific Reports, 7(1). doi:10.1038/s41598-017-13608-5
Hendler, R. W., Barnett, S. M., Dracheva, S., Bose, S., & Levin, I. W. (2003). Purple membrane lipid control of bacteriorhodopsin conformational flexibility and photocycle activity. An infrared spectroscopic study. European Journal of Biochemistry, 270(9), 1920-1925. doi:10.1046/j.1432-1033.2003.03547.x
Back to Top