Viperin: A putative radical SAM enzyme
Jeremy Moore '19 and Sasha Smerakanych '20
Contents:
I. Introduction
To respond efficiently to infection, cells produce signaling proteins called interferons (IFNs). By inducing expression of IFN-stimulated genes (ISGs), these proteins are capable of activating several antiviral processes, including apoptosis, and viral RNA destruction. One common ISG product is viperin, an enzyme induced by IFN-α and IFN-β which is able to inhibit viral replication. Localized in the endoplasmic reticulum, it has been discovered to impede a wide range of viruses, including the human cytomegalovirus, HIV-1, West Nile, hepatitis C, influenza A and tick-borne encephalitis. Despite our knowledge of its biological function, the details behind viperin's enzyme mechanism are not understood.
Viperin's amino acid sequence contains a CxxxCxxC motif, which is characteristic of radical S-adenosylmethionine (SAM) enzymes, a protein superfamily that produces a free radical by splitting SAM using a [4Fe-4S]
.
These enzymes then use this radical to conduct additional chemistry.
Viperin is highly conserved in vertebrates and several homologues exist in fungi, bacteria and archaea. To understand how this enzyme works, and see whether it could actually be a radical SAM enzyme, Fenwick et al crystallized an N-terminal truncated viperin with the SAM analog, S-adenosylhomocystein (). Using this crystal structure, they were able to show that the structure contains a canonical SAM-enzyme active site, and aligns well with enzymes that catalize similar reactions.
II. General Structure
Viperin is a globular protein that contains a partial (βα)6-barrel from residues 75 to 284. This fold appears often in radical SAM enzymes.
The partial (βα)6-barrel is augmented by an additional short (residues 290 to 293), a (residues 304 to 312) , and three (residues 295 to 301 and 325 to 336), which together make up a region called the .
The C-terminal extension (residues 290-336) folds over the (αβ)6-barrel. Residues 337-362 of this extension are predicted to be disordered.
Although viperin was crystallized without the C-terminus, this region is crucial for Viperin's enzymatic activity. A conserved tryptophan in this region has been shown to interact with the Fe/S cluster assembly factor CIAO1, without which, the iron sulfur cluster will not be incorporated into Viperin. Thus, in-vivo, a truncated viperin would be enzymatically inactive.
Viperin topology
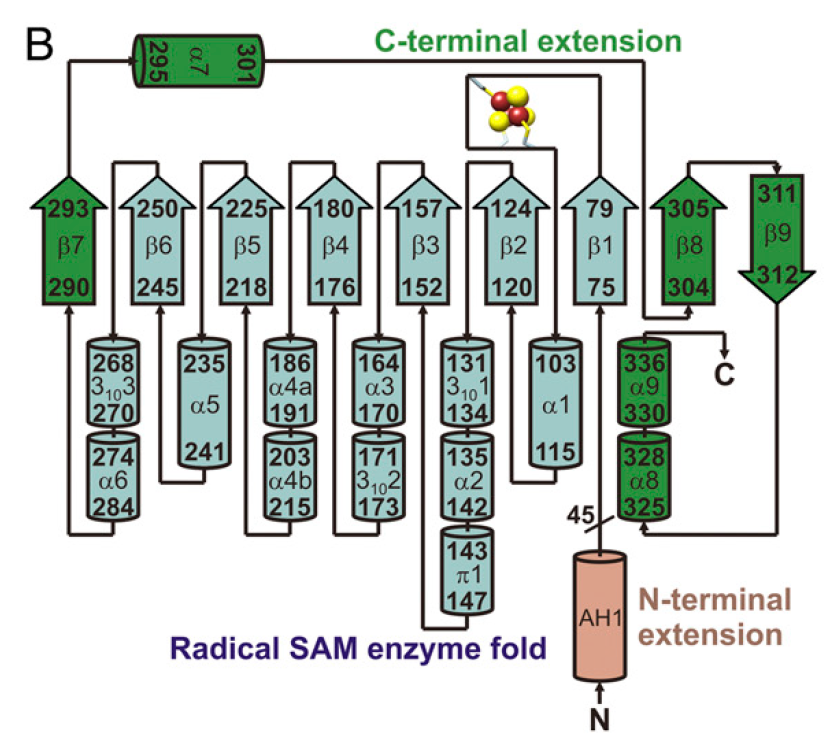 |
III. Radical SAM Enzyme Etructure
The (αβ)6-barrel fold in viperin contains a
from residuces 84 to 91. Here the cysteine residues that coordinate the iron-sulfur cluster are shown in
purple. These residues orient the single free iron towards the active site, where it is able to interact with SAH. Viperin's interactions with the iron sulfur cluster and substrate are consistent with those of known SAM-enzymes.
Hydrogen bonds between the α-amino and α-carboxy groups of SAH and the un-ligated iron of the [4Fe-4S] cluster the substrate in the active site. Additionally, in the active site, the α-carboxylate group on SAH can hydrogen bond with Arg194, and Ser180.
There are also stabilizing hydrogen bonds between the 2' hydroxyl on SAH and Arg194. Similarly, the 3' hydroxyl forms hydrogen bonds with Ser180 and Asn222.
While the back end of SAH is stabilized by hydrogen bonds, the adenine moiety is primarily stabilized by hydrophobic interactions with
Phe92,
Phe90, and
Val224.
In addition to hydrophobic interactions, the adenine moeity interacts with a polar site near the (displayed as a cartoon). This loop also binds the [4Fe-4S] cluster, which helps position it by the substrate.
IV. Active Site Channel
A 6 angstrom leads to the C5' of SAH, where radical formation likely occurs. This channel is made by both the (αβ)6-barrel and the C-terminal extension. A stems from the opposite side of the protein, formed by Leu264, Arg265 and the cluster binding loop.
The is formed by residues from β1-β6 as well as the first few residues of the C-terminal extension. In addition to several uncharged residues, The active site contains five basic (Lys120, Lys220, Arg245, Lys247, and Lys297), and one acidic residue (Glu293).
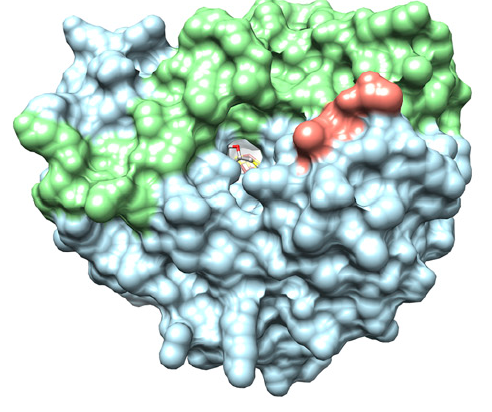 |
| Solvent accessible surface of Viperin |
V. Comparison with other SAM enzymes
The putative active site residues in Viperin are highly conserved among vertebrates. Viperin also appears to have some close fungal relatives that lack the N-terminal extension, but have a C-terminal extension that may perform the same function.
While the function of Viperin homologues in fungi is unknown, their sequence similarity suggests that they catalyze similar reactions.
According to a Dali search, Viperin and MoaA, another radical SAM enzyme, appear to have highly similar structures. MoaA is a protein found in both prokaryotes and eukaryotes involved in molybdopterin biosynthesis. Superposing these structures shows that the partial (βα)6-barrel cores overlap well. Additionally, both enzymes coat this barell with charged residues.
Unlike the
viperin C-terminal extension, the
MoaA C-terminal extension ligates another [4Fe-4S] cluster, but is still placed in a similar location. Despite accepting slightly different substrates, the active site similarities between MoaA and Viperin present additional evidence
that Viperin performs a radical-based reaction.
VI. References
Broderick JB, Duffus BR, Duschene KS, Shepard EM. Radical S-adenosylmethionine enzymes. 2014. Chem Rev, 114:4229-42317.
Fenwick MK, Li Y, Cresswell P, Modis Yorgo, Ealick SE. Structural studies of viperin, an antiviral radical SAM enzyme. 2017. PNAS, 114(26):6806-6811.
Hanzelmann P, Schindelin H. Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. 2005. PNAS, 103(18):6829-6834.
Back to Top