U6 snRNP in complex with Prp24
Aidan Ohning '20 and Alex Law '20
Contents:
I. Introduction
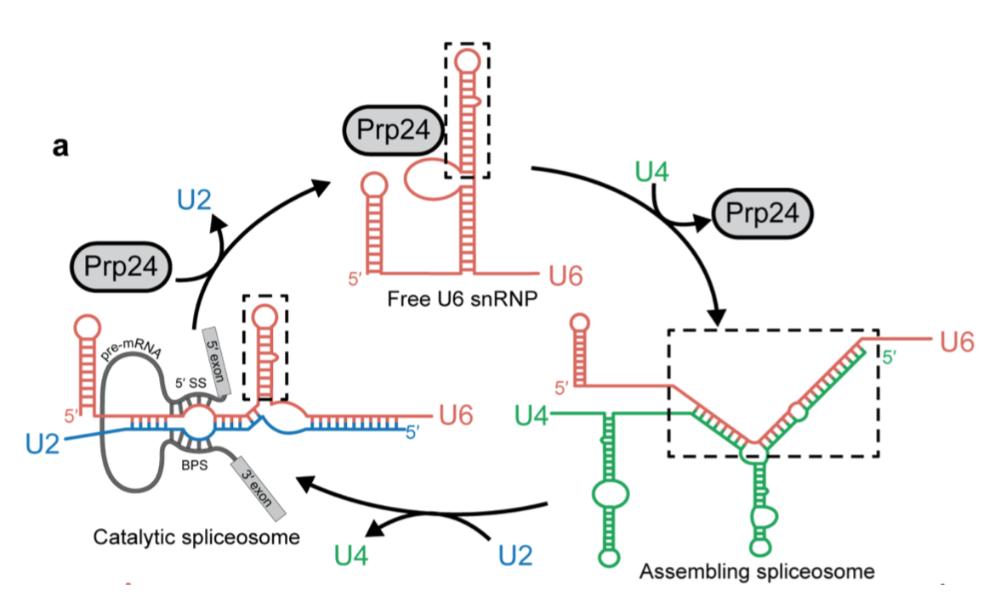
The spliceosome contains five small nuclear ribonucleoproteins
(snRNPs), working to remove introns from pre-mRNA. The snRNPs are
named U1, U2, U4, U5 and U6, the “U” is due to the high
concentration of uridine within them. Spliceosome assembly occurs
due to the ordered interactions of the spliceosomal snRNPs.
U1 binds to the 5’ end of the pre-mRNA splice site to
initiate the spliceosome assembly. U2 binds to an adenosine with a
free 2’ hydroxyl group. ATP is hydrolysed to bring U1 and U2
together. U4, U5, and U6 bind with U1 and U2, bringing them closer
together to bring the 2’ hydroxyl closer to the 5’ end of the
intron. U1 and U4 are released via ATP hydrolysis. A lariat intron
is formed and released along with the snRNPs. At this point the
two ends of the exon are ligated together.
. 
snRNPs are the most conserved small nuclear RNAs and are
important players in catalysis. Together U6 and Prp24 may nucleate
the annealing of U4 and U6 snRNPs. This is because there are four
RNA Recognition Motif (RRM) domains within the Prp24 protein.
1, 2, and 4 help form
an electropositive groove that binds dsRNA, causing the nucleation
of annealing.
II. U6-Prp24 Binding Motifs
The U6-Prp24 structure confirms the existence of the
proposed telestem region in U6. The telestem and the ISL are
nearly perpendicular to one another, they are separated by a
nucleotide asymmetric internal loop. This RNA loop forms an
interface with RRMs 2 and 3. This interface between the RRMs
contains a highly distorted conformation of RNA with a
motif.
The ‘skip-stack turn’ motif is located next to the 5’ splice site
binding region in U6. It is a novel motif similar to the Z-turn motif.
Skip-stack turn contains alternating stacked bases which are stabilized by the
RNA and residues found within the N terminal of RRM1. RRM1 contacts RNA within
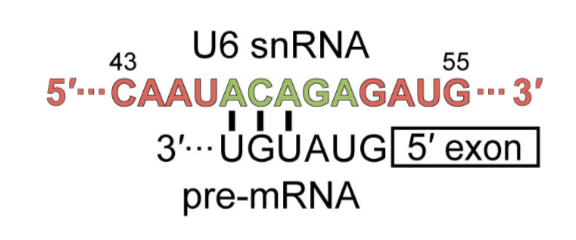
its neighboring complex. RRM2 interacts with the highly conserved ‘ACAGA’ box,
which then binds to the 5’ splice site in the assembled spliceosome.
RRM2
interacts with the highly conserved
box, which then
binds to the 5’ splice site in the assembled spliceosome.
. 
The
is
another novel motif which bulges two vicinal nucleotides to
permit stacking of A56 and A59. The stacked nucleotides -- in
conjunction with 3’ end of the “Skip-Stack-Turn” -- aid in the
formation of a hydrophobic cage around Phe134 of RRM2. This
helps mediate
tertiary interactions between RRMs 2 and 4.
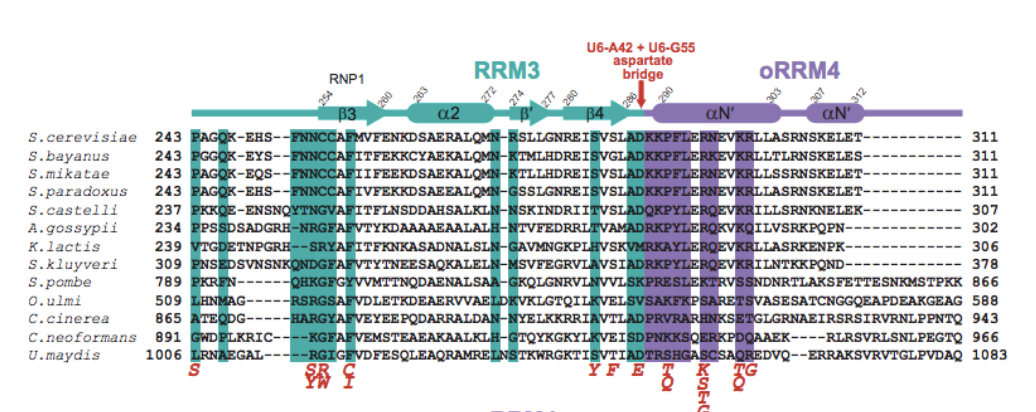
An "Aspartate bridge" is formed via Asp288 hydrogen
bonding with both A42 and G55 on opposing sides of the
asymmetric bulge, linking protein and RNA. This
helps further
stabilize tertiary interactions between the RRMs.
III. U6 Mutations
Stable interactions between Prp24 and U6 is in conflict
with its role in the annealing of U4/U6. The splicing cycle is made up of
numerous equilibria, causing high temperature sensitivity. This temperature
sensitivity is a valuable tool for investigating the RNA base-pairing
dynamics in vivo. In the Prp24-U6 complex there are 32 trans-acting
suppressors, these reduced cold sensitivity. The A62U C85A double mutation
within U6 allowed for cold resistant growth at 30° C. Many similar mutations
lie on the RNA-protein interface, influencing the U6 snRNA equilibria.
However, the mechanisms of these suppressors is poorly understood, most likely
working through destabilization of the U6 Prp24 complex.
All four of the components of the aspartate bridge are sites of substitution.
Positioning of the side chain carboxylate group and backbone is integral to the
structural integrity of the bridge. At the telestem-asymmetric bulge junction there
are also suppressor mutations, affecting the hydrogen bonding network.
Mutations at , ,
and suppress cold sensitivity. These factors
indicate that destabilization at multiple suppressor sites can compensate for the
hyperstabilization of the ISL. Investigations into these suppressors show the possibility
of reducing sensitivity within other spliceosome equilibria.
. 
. 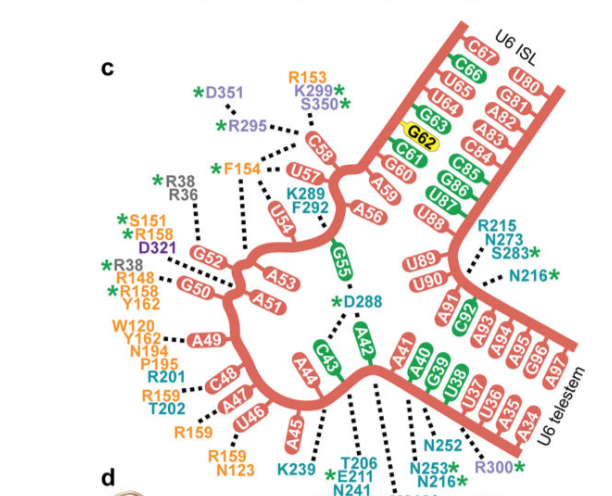
animated from figure (d) illustrated directly above.
VI. References
Eric J Montemayor, Elizabeth C Curran,
Hong Hong Liao, Kristie L Andrews, Christine N Treba, Samuel E Butcher,
David A Brow. Core structure of the U6 small nuclear
ribonucleoprotein at 1.7-Ĺ resolution.Nature Structural and Molecular Biology,
2014; DOI: 10.1038/nsmb.2832
Will CL, Luhrmann R. Spliceosome structure and function.
Cold Spring Harb Perspect Biol.
(2011) 3:a003707. 10.1101/cshperspect.a003707
Wan, R.; Yan, C.; Bai, R.; Wang, L.; Huang, M.; Wong, C. C. L.; Shi, Y.
3.8 A Structure of the U4/U5 U6 tri-snRNP: Insights into spliceosome assembly and catalysis.
Science 2016, 351(6272), 466–475.
Back to Top