Rad51 Filament as a DNA
Homologous Recombinase
Sofia Rehrig 23' and Erika Conant 23'
Contents:
I. Introduction
Homologous recombination is a highly reserved
process in both eukaryotic and prokaryotic organisms, effective in the
elimination of deleterious lesions, including double-stranded breaks
and interstrand crosslinks from chromosomes, as well as the
preservation of replication forks, telomere maintenance, and
chromosome segregation in meiosis I. Rad51 serves as a catalytic core
of homologous recombination for eukaryotes via through a filamentous
intermediate on the ssDNA, called the presynaptic filament. This
results from Rad51s ability to repair double-stranded breaks in DNA,
as well as its capacity to provoke genetic diversity.
In this study, we delve into the structure and
mechanistic pathway of the Rad51 filament, as well as a mutant,
pertaining to His352, which has been crystalized in Saccharomyces
cerevisiae. In order to understand the implications of Rad51, it may
be helpful to knwo the general mechanism of the process, as presented
in Figure 1. This diagram clearly outlines the ways in which
synthesis-dependent strand-annealing (SDSA) and double-strand break
repair (DSBR) function to produce crossover or noncrossover products.
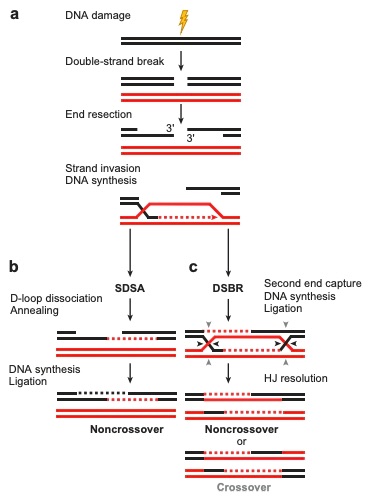
Figure 1. A general mechanism of
homologous recombination in both the SDSA and DSBR pathways. (Filippo,
J.S., Sung, P., Klein, H., 2008)
II. General Structure
Rad51 is composed of a series of
(labeled A-G) that encase ssDNA to form the presynaptic
filament. Within each promoter is a
that contains a central core surrounded by four
alpha-helicies, and a fifth alpha-helix
that is separate from the central four and projected N terminally.
There are also
within each promoter, and these are responsible for promoter-promoter interactions.
The molecule to the left is arranged as it would be when bound to ssDNA (ssDNA not shown). Interactions with ssDNA change the helical pitch and conformation of Rad51, and
Rad51-ssDNA has a helical picth of 103A, comprising 6.4 promoters per turn, with a rise of 16.1A and a twist of 56.2 degrees.
Additionally, Rad51-ssDNA interactions have effects on inter-promoter distance, with shorter distances reported for Rad51 not bound to ssDNA.
Despite the differences noted above, electron microscopy has shown that the promoter-promoter interfaces
within each Rad51 structure are invariant.
III. Mechanistic Approach of Homologous Recombination
Homologous recombination begins with the 5 to
3 endonucleolytic resection of DNA, resulting in a tail. Replication
Protein A (RPA) momentarily interacts with the tail, but is soon
replaced by ATP-bound Rad51, which coats the tail to produce the
presynaptic nucleoprotein filament with the ssDNA attaching at
and
This interaction is stabilized through the BRC repeat motif
with the BRCA2 tumor suppressor protein. It is then able to catalyze
the exchange of strands between the ssDNA and dsDNA substrates,
initiating strand synthesis and error-free repair. ATP hydrolysis by
the catalytic activity of RAD51 allows the components of the repair
reaction to dissociate." 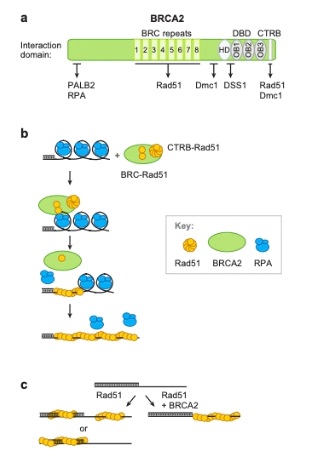
Figure 2. Mechanism of Homologous
Recombination, shown using functional domains of human BRCA2
recombination mediator activity. (Filippo, J.S., Sung, P., Klein,
H., 2008)
IV. Mutation of His352
Much similar to the human version of Rad51, the
yeast homolog found in Saccharomyces cerevisiae, holds a common
function within homologous recombination. Conway et al. delineated
that this yeast homolog contains a mutation of the
which lies at the protein-protein interfaces, and markedly
disrupts DNA binding. It is believed that this particular histidine is
involved in the catalysis of ATP hydrolysis and most likely plays an
important role in allosteric regulation of the system.
The conformation of the mutated filament can
participate in promoter-promoter interactions with
at the
. The ATPase site site is
composed of three different amino acids that form a promoter-promoter
Interface.
can be phosphorylated by c-abl, and is able to stack against
(
which provides aromatic stability to the structure) from an
adjacent promoter's ATPase domain, and
helps to anchor the trans beta-strand onto the adjacent
promoter. There are two ways that the promoter-promoter interactions
can occur. In the first, His352
from one promoter is positioned directly over the
ATPase site of the other. In the second interaction, the
helix bearing the His352 twists 12° relative to the C-terminal domain
and 5° relative to the adjacent C-terminal domain, moving it away from
the ATPase site. Because of this,
His352is sterically occluded from the
ATPase site by Phe187 of
the adjacent promoter.
This gain-of-function mutant has a longer pitch
than that of the prokaryotic homolog, RecA (73-83 Ε), reaching 130 Ε.
Studies have also indicated that this mutant is capable of binding 4±1
nucleotides per promoter. This data gives reasoning to the
, as well as raising the question of the stoichiometry at play.
V. References
Bonilla, B., Hengel, S. R., Grundy, M. K.,
and Bernstein, K. A. (2020). RAD51 Gene Family Structure and
Function. Annual Review Genetics. 54: 2546. doi:
10.1146/annurev-genet-021920-092410. Conway, A. B., Lynch, T. W., Zhang, Y., Fortin, G. S.,
Fung, C. W., Symington, L. S., and Rice, P. A. 2004. Crystal
structure of a Rad51 filament Nature structural and molecular
biology, 11(8), 791-796. https://doi.org/10.1038/nsmb75
Filippo, J. S., Sung, P., and Klein, H.
(2008). Mechanism of Eukaryotic Homologous Recombination. Annual
Review Biochemistry, 77:22957. Doi:
10.1146/annurev.biochem.77.061306.125255
Ristic, D., Modesti, M., van der Heijden,
T., van Noort, J., Dekker, C., Kanaar, R., Wyman, C. (2005). Human
Rad51 filaments on double- and single-stranded DNA: correlating
regular and irregular forms with recombination function. Nucleic
Acids Research. 33(10):3292-302. doi: 10.1093/nar/gki640.
Print 2005.
Short, J. M., Liu, Y., Chen, S., Soni, N.,
Madhusudhan, M.S., Shivji, M. K. K., and Venkitaraman, A. R.
(2016). High-resolution structure of the presynaptic RAD51
filament on single-stranded DNA by electron cryo-microscopy. Nucleic
Acids Research. 44(19), 90179030. doi: 10.1093/nar/gkw783.
Back to Top