A. Thaliana Rubisco
Brent Henderson '22 and Jack Caine '24
Contents:
I. Introduction
Rubisco, ribulose-1,5-bisphosphate carboxylase-oxygenase, is an
enzyme found in all autotrophs and has been deemed the most abundant
enzyme on the planet. Rubisco acts primarily to fix atmospheric CO2.
Rubisco catalyzes the first step of the Calvin cycle which takes up
atmospheric CO2 and sees it interact with RuBP(ribulose bisphosphate)
to form an organic compound that can continue the rest of the Calvin
cycle. However, CO2 and O2 compete for the binding domain of rubisco.
This competition drastically reduces the efficiency of the enzyme
which necessitates its presence in large quantities to ensure enough
CO2 is available to keep the biological rate of reaction high enough
for the plant to have enough energy.
Rubisco is an interesting enzyme to study because of its high
abundance, low efficiency and its essential role within the Calvin
cycle. More specifically in Arabidopsis Thaliana (A. Thaliana),
the enzyme structure has only recently been characterized. A.
Thaliana is a model organism for higher plants, so
characterizing its rubisco could help to gain insight into the
structure and function of rubisco in higher plants. Other autotrophs
like moss, algae and bacteria that are not categorized as higher
plants still contain rubisco and can benefit from the study in A.
Thaliana.
The structure and efficiency of rubisco have dramatic
implications for the rate of CO2 uptake as well as overall plant
growth. One potential goal for understanding Rubisco is to develop a
similar enzyme that could more efficiently intake CO2. This could
have extreme benefits on plant growth and on enviornmental CO2
levels.
II. General Structure
This crystal structure of this Rubisco is one quarter of its
full hexadecamer structure(Figure 1). A full A. Thaliana
Rubisco has eight large (LSu ) and
eight small (SSu) subunits, with active
sites present between large subunits. Thus, A. Thaliana
rubisco has eight active sites in nature to increase its rate of
oxygenase or carboxylase.
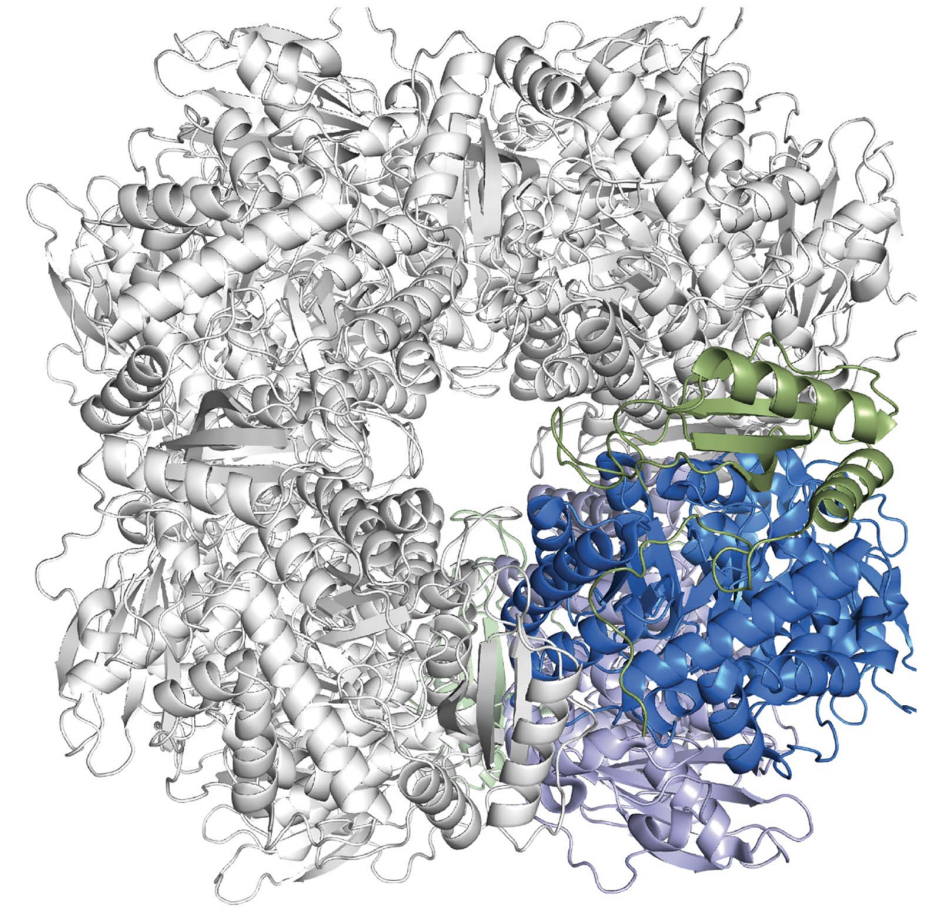
Figure 1: A representation of the hexadecamer; this is the full
strucutre of Rubisco. The crytal stucture used in this tutorial is
shaded in color (Valegard et al.).
The crystal structure used in this tutorial is a quarter,
having two large subunits and
two small subunits; and therefore one active site.
In addition to the L2S2 subunits present, the crystal
structure was bound to a 2-carboxyarabinitol-1,5-bisphosphate
(2-CABP), a transition state analogue. This crystal structure
represents the active state of the enzyme.
III. Large LSu subunit
The Rubisco large subunit, deemed LSu
, is composed of 479 amino acids and is mainly responsible
for the catalytic activity. Catalysisoccurs between two LSu
subunits. There are 2 LSu
subunits in this crytal strucutre.
A single LSu subunit
consists of a N-terminal domain ,
residues 1-150, and a C-terminal domain,
residues 151-479. The N-terminal domain
consists of 4 Beta-sheets and 2 alpha-helices. The C-terminal
domain contains 8 individual Betaalpha-barrels.
The active site lies between the two LSu
subunits. The N-terminal domain
has 4 catalytic residues: Tyr20,
Glu60, Thr65 and Asn123. These key 4 residues of the N-terminal
active site connect with C-terminal
residues in the adjacent LSu
subunit: Lys175,
Lys177, Lys201, Asp203, Glu204, His294, Arg295, His327, Lys334 and
Leu335. Also of importance, a Mg2+
ion helps stabilize the Lys201
residue.
IV. 2-carboxyarabinitol- 1,5-bisphosphate Binding
2-carboxyarabinitol-1,5-bisphosphate
(2-CABP) is the synthetic molecule that binds to the active
site between LSu subunits.
2-CABP binds nearly irreversibly and in a stoichimetric
manner. 2-CABP is related to
2-carboxyarabinitol-1,5-bisphosphate, a natural inhibitor of
Rubisco. The 2-CABP is not shown in
the crystal structure, but was emphasized in this tutorial as it is
an example of how a subtrate binds to the active site of Rubisco
(figure 2). Furthermore, the 2-CABP
was necessary for the crystal structure to be produced.
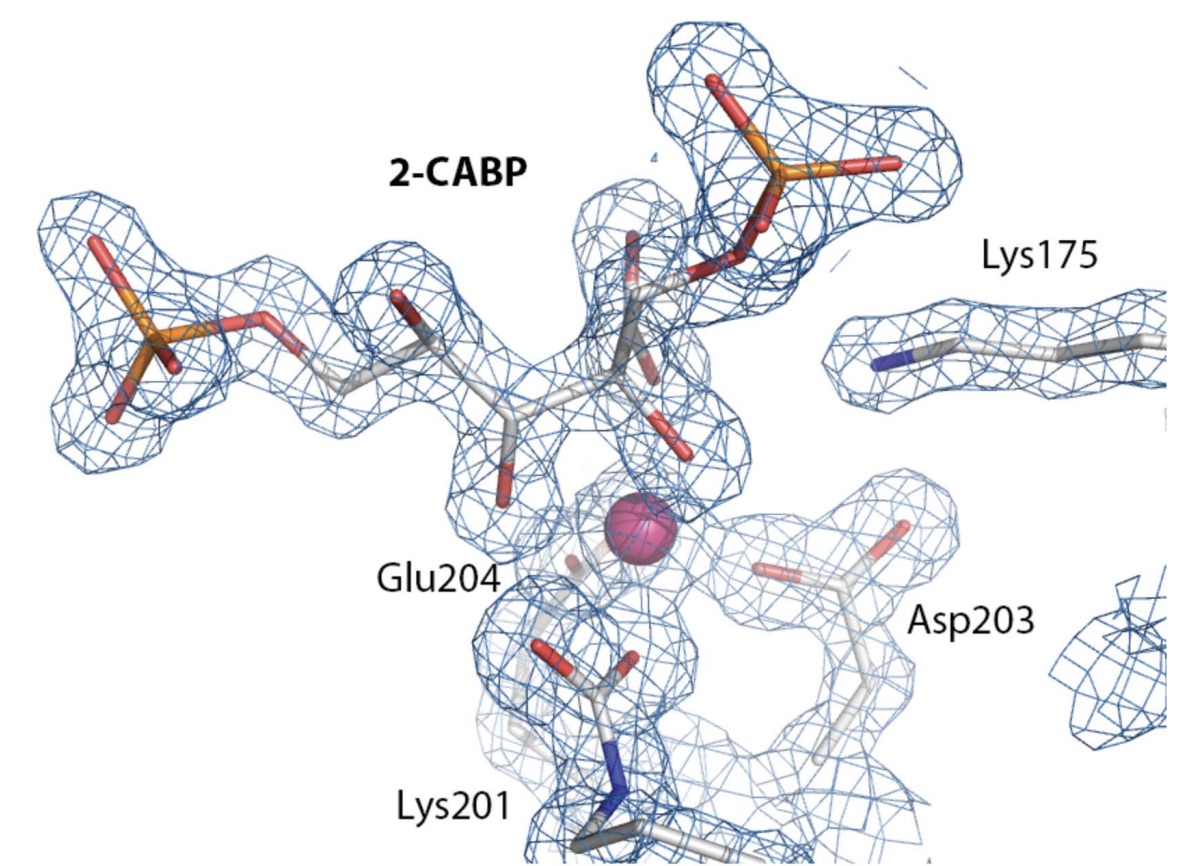
Figure 2: 2-CABP bound to the active site in Rubisco. This helped the
researchers gather the crystal structure of Rubisco. The Mg ion that
stabilizes the binding is shown in pink (
Valegard et al.).
V. Small SSu Subunit
The Rubisco small subunit, SSu,
lies at the core of the enzyme. SSu
consists of 125 amino acids.
This small subunit consists of 4 beta-sheet strands and 2
alpha-helices, similar to the N-terminal domain of LSu
.
The key characteristic of SSu
is a betaA-betaB loop, which is 22
residues long
. This extends into the solvent channel of the Rubisco
protein.
There exist multiple different isoforms of the SSu
subunit that exists within the L8S8 Rubisco structure in A.
thaliana. The strucutre presented here is the RbcS1B SSu
isoform. This differs from the RbcS1A SSu
isoform at amino acid residues 2, 24, 34,
58, and 96. A. thaliana Rubisco SSu
isoforms Rbcs2B and RbcS3B are identical, and only differ from the
RbcS1A SSu isoform shown here at residue
22.
VI. References
Andersson, I., Backlund, A. Structure and
function of Rubisco. Plant Physiology and Biochemistry, vol.
46, 2008, pp. 275-291.
Valegard, Karin. Structure of Rubisco from
Arabidopsis Thaliana in Complex with
2-Carboxyarabinitol-1,5-BisPhosphate. Structural Biology,
vol. 74, part 1, Jan. 2018, pp. 1-9.
http://scripts.iucr.org/cgi-bin/paper?S2059798317017132\ Science
253: 1001-1007.
Fristedt, Rikard. RAF2 Is a RuBisCO Assembly
Factor in Arabidopsis Thaliana. Plant Journal, vol. 94,
no. 1, 3 Feb. 2018, pp. 146-156. https://doi.org/10.1111/tpj.13849
Servaites, J C. Inhibition of ribulose
1,5-bisphosphate carboxylase/oxygenase by
2-carboxyarabinitol-1-phosphate. Plant physiology vol.
92,4 (1990): 867-70. doi:10.1104/pp.92.4.867
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1062388/?page=1
Back to Top