Mycobacterium tuberculosis
response regulator PhoP
Darya Aminia '25 and Minh Pham '23
Contents:
I. Introduction
PhoP belongs to the OmpR/PhoB transcription regulator
subfamily, which is conserved in both Gram-negative and Mycobacterium
pathogens and is important for bacterial virulence. The full
structure of PhoP has been reported for Mycobacterium
tuberculosis (MTB), which is responsible for the
highly infectious tuberculosis disease. In MTB, PhoP transcription
regulator regulates more than 110 genes and plays an important role
in the bacterium's virulence. It is a protein with 250 amino acids.
Unactivated PhoP exists as free monomers. Phosphorylation of
the N-terminal receiver domain in PhoP by an upstream histidine
kinase causes the activated proteins to form compact
that bind two direct repeated
The cooperative binding of PhoP in oligomers (most commonly
dimers) to the direct repeat sequences in MTB promoters upregulates
transcription of target genes.
II. General Structure
PhoP is about 30kDa in size, consisting of only 1 polypeptide
chain. PhoP, like all RRs of the OmpR/PhoB subfamily, consists of
two well-structured N-terminal signal
(RD) and C-terminal
(DBD) linked together by a short, disordered
(9 amino acids).
Both RD and DBD consist of
and
with hydrophobic interactions making up the bulk of
intermolecular interactions inside each domain. PhoP is loose
and flexible in non-binding, unactivated form (free monomer) and
compact in DNA-binding, activated form (tandem dimer). In
unactivated PhoP, RD and DBD do not contact each
other, but they do
in the activated DNA-binding form.
III. Reciever Domain
The N-terminal receiver domain has a (??)5-fold structure,
where a hydrophobic five-stranded parallel ?-sheet
is surrounded by amphiphilic
? 1 and ? 5 on one face and helices ? 2, ? 3, and ? 4
on the other face. The majority of hydrophobic residues in the core
?-sheet are Val, Leu, and Ile. The inactive RDs can form
with each other via interactions between their ? 4-?
5-? interfaces. However, these inactive dimers are two-fold
symmetrical not tandem, rarely occur, and have no transcription
regulating activity.
The activation pocket lies at the
and is lined with all
apart from a well-conserved basic
residue (He, 2016). This acidic pocket contains the
phosphoryl acceptor site
that is conserved in all OmpR/PhoB transcription
regulators.
Activated RD has been crystallized with
. BeF3 can be seen as the proxy for the additional
negatively charged oxygen on the phosphorylated Asp71
residue. The phosphorylated Asp71
would be stabilized with: 2 hydrogen bonds with Lys121
and Leu72 backbone and 2 charged interactions with a Ca2+ ion. The
is suspended in a hexa-coordination by interactions
with Asp27,
Glu29, Met73, and Lys121.
The structures of activated and unactivated receiver domains
are almost identical (Figure 1), and the receiver domains do
not ever interact with DNA. Thus, little is known about the
activation mechanism of PhoP after phosphorylation at the conserved
Asp71.
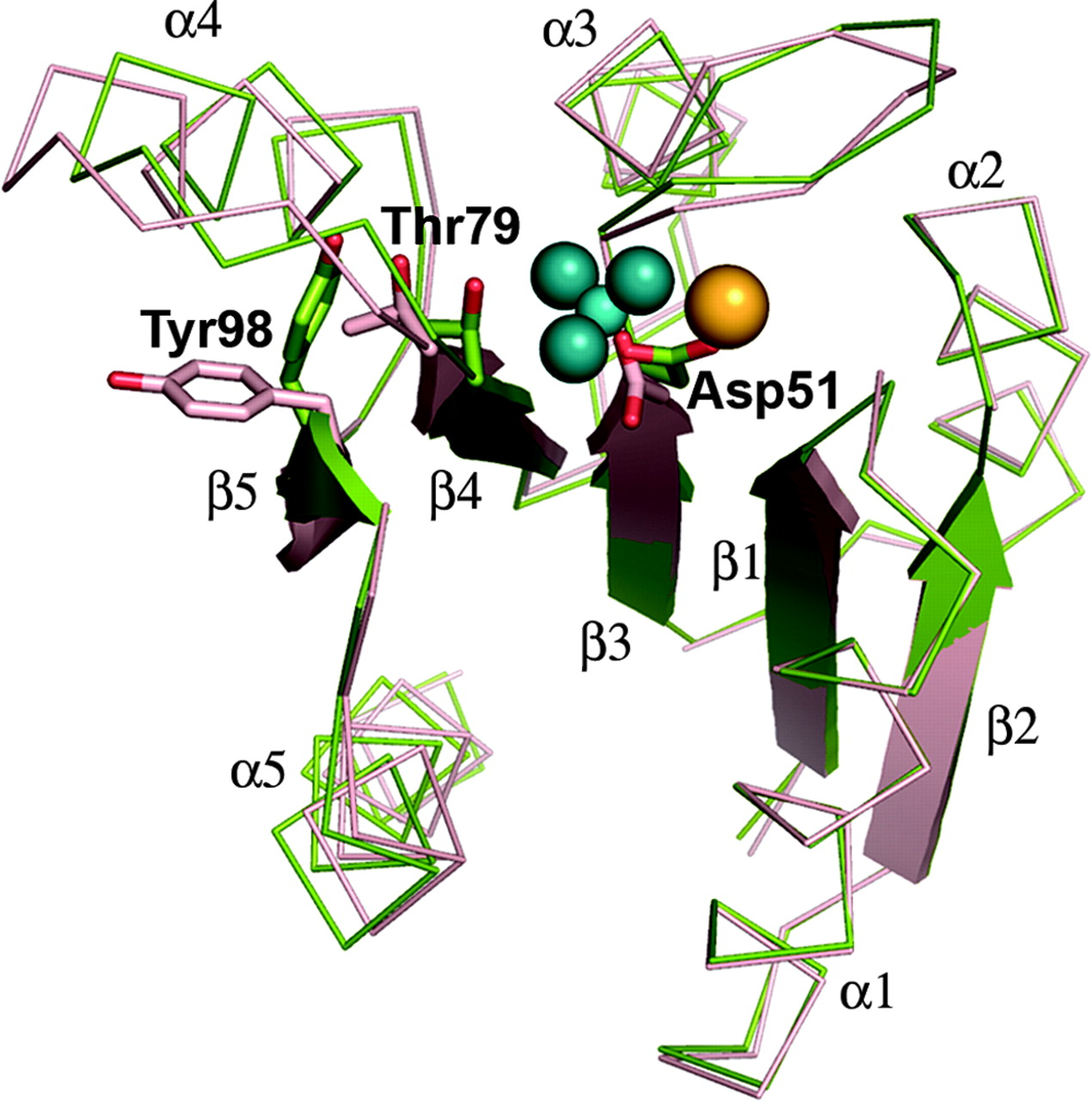 Figure 1: Overlay of activated and unactivated PhoP
receiver domains. Reproduced from Bachhawat and Stock, 2007.
Figure 1: Overlay of activated and unactivated PhoP
receiver domains. Reproduced from Bachhawat and Stock, 2007.
IV. DNA Binding domain
The C-terminal DNA-binding (effector) domain has a winged
helix-turn-helix (HTH) structure typical of DNA-binding
proteins. It starts with a 4-stranded antiparallel ?-sheet, followed
by three perpendicular ?-helices and then a C-terminal ?-hairpin.
The ?-sheet and ?-hairpin are the
structure.
PhoP effector domain has a positively charged surfacethat is
complimentary to the negatively charged DNA backbone.
Sequence-specific interaction between the PhoP effector domain and
its target TCACAGC
motif is facilitated by aromatic and hydrophobic side chains on or
near the alpha-8 helix on the effector domain.
The
invades the major groove of the target TCACAGC motif and uses
the residues Asp212,
Tyr217, Ser216, and Glu215 to
form
with the DNA bases.
from the beta-hairpin also forms sequence-specific and backbone
interactions in the adjacent downstream minor groove from the
alpha8-binding site.
PhoP effector domain only binds DNA in
or higher plurality. Each effecter domain from a
subunit of PhoP tandem oligomers bind one TCACAGC motif in a series
of direct repeats (most commonly 2) (Figure 2) separated by a
strict
.
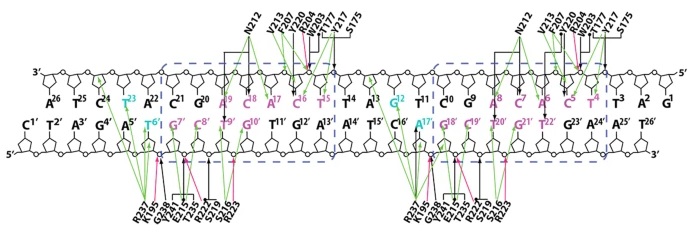 Figure 2: Detailed scheme of interactions between
effector domains in PhoP tandem dimers and DNA at the TCACAGC direct
repeats. Interactions of both subunits in tandem primers are the same
and are both shown in this diagram. This figure is reproduced from He
et al., 2016.
Figure 2: Detailed scheme of interactions between
effector domains in PhoP tandem dimers and DNA at the TCACAGC direct
repeats. Interactions of both subunits in tandem primers are the same
and are both shown in this diagram. This figure is reproduced from He
et al., 2016.
V. Tandem Dimers
Two subunits of the tandem primer have the same orientations
and different environments. Hence, they are not interchangeable like
subunits of symmetric dimers.The PhoP molecule in the tandem dimer
that binds to the first TCACAGC motif in the direct DNA repeat is
referred to as A and the molecule that binds to the second motif
downstream is referred to as B. Both
in the dimer are expansive.
The intersubunit interactions are divided into the major
patch and the minor patch. The
includes helices ? A, ? A, ? 3 B, and ? 4 B from RD A, DBD A, and
RD B. Helix ? 4 is highly flexible. Its different conformations are
the key distinctions between the three states of PhoP:
is a one-turn helix.
is a 1.5-turn helix.
- which lies at the center of the major patch, is unwound.
Residue
of ? 4 B buries into a shallow hydrophobic pocket of RD A. The
unwound ? 4 B also forms a hydrogen bond, hydrophobic interactions,
and a pi-pi stack with
on alpha7 A. Interactions between
in the major patch are primarily mediated by Arg84 and Arg87 on
? 3 B and Glu34 on ? 1 A.
The
occurs between two DBDs. The C-terminal ?-hairpin A from DBD A
and the loop between ? 7 B and ? 8 B strands of DBD B form
hydrophobic backbone and residue interactions *button*, a hydrogen
bond (Glu161 A - Val192 B) *button*, and a charge interaction
(Glu164 A - Arg244 B) *button* with each other.
The
in the tandem dimer determines PhoP's efficacy in transcription
induction. Mutations in the RD A hydrophobic pocket (Tyr 205) or
Leu113 residue of the unwound ? 4 B significantly decreases PhoP
dimers' ability to induce transcription. Additionally, the
ensures activated PhoP subunits stay on the same side of the
DNA helix and achieve optimal intersubunit interactions. So,
mutations that change the spacer sizes of PhoP targets heavily
punish transcription induction. This explains PhoP's preference to
bind DNA in tandem dimers and oligomers.
VI. References
Menon, S., Wang, S. (2011). Structure of the
response regulator PhoP from Mycobacterium tuberculosis reveals a
dimer through the receiver domain.Biochemistry, 50(26),
5948-5957.
Bachhawat, P., & Stock, A. M. (2007).
Crystal structures of the receiver domain of the response
regulator PhoP from Escherichia coli in the absence and presence
of the phosphoryl analog beryllium fluoride. Journal of
bacteriology, 189(16), 5987-5995.
Wang, S., Engohang-Ndong, J., & Smith, I.
(2007). Structure of the DNA-binding domain of the response
regulator PhoP from Mycobacterium tuberculosis. iochemistry,
46(51), 14751-14761.
He, X., Wang, L., & Wang, S. (2016).
Structural basis of DNA sequence recognition by the response
regulator PhoP in Mycobacterium tuberculosis. Scientific
reports, 6(1), 1-11.
Macdonald, R., Sarkar, D., Amer, B. R., &
Clubb, R. T. (2015). Solution structure of the PhoP DNA-binding
domain from Mycobacterium tuberculosis. Journal of
biomolecular NMR, 63(1), 111-117..
Back to Top