A COVID-19 Accessory Protein, SARS-CoV-2 ORF3a
Colin Shin '24 Rachel Chen '24
Contents:
I. Introduction
The novelly discovered SARS-CoV-2-encoded ORF3a is of high interest to
researchers combatting the COVID-19 pandemic. SARS-CoV-2 ORF (open
reading frame) 3a encodes a highly conserved, putative viroporin across
SARS-Cov-2 variants. Viroporins are transmembrane channels that alter
the infected cell's permeability to allow entry of permeant ions and
small molecules, which can cause a plethora of homeostatic disruptions
to the cell and facilitate viral release, which drastically enhances
viral growth of SARS-CoV-2 (Breitinger, et al.). Viroporins have also
been found to trigger inflammatory responses and suppress defensive
apoptosis of coronavirus-infected cells (Ren, et al.). Based on Cryo-EM
data, we can now analyze ORF3a's structure for potential insights on how
it directs viral-host interactions.
II. Structural Overview
Starting at the N-terminus, the SARS-CoV-2 3a protomer has a
transmembrane region,
consisting of 3 alpha helices (TM1,TM2,TM3),
connected to a
cytosolic domain (CD) by a
turn-helix-turn
(1). The
protrudes the viroporin into cytosol. It contains 8 beta strands in a
commonly observed beta-sandwich, where pairs of antiparallel beta
strands stack atop each other. B1, B2, B6, and partially B7 make the
"outer" sheet; while, B3, B4, B5, B8, and half of B7 make the "inner"
sheet. This is a novel fold that has not yet been observed in other
proteins.
Kern, et al. has successfully reconstituted SARS-CoV-2 3a as dimers
and tetramers, but the molecule could potentially oligomerize even
larger. Dimeric contact is facilitated by highly complementary
interactions between the CD using these specific
(Kern et al.). This results in a hydrophobic core that
buries up to 940 angstrom-squared of surface area per beta chain. The
lumenal side will show TM1-3 of one protomer and TM1-3 of another
respectively encircled in a
manner.
The 124 kDa tetramer is a side-by-side arrangement of two identical
dimers (Figure 1). Cyro-EM analysis suggests this contact to occur
between the TM3-CD linker and the beta1-beta2 linkers of neighboring
dimers, at the
This small contact between loops buries only 600
angstroms-squared of protein surface. Consistently, the tetramer has
been less frequently observed than the dimer. Experimentally
introduced mutations to a
adjacent below TM3 but above beta1 and beta2 propound that
disulfide bonding also facilitates tetramerization. The bonds most
likely occurs between
, whose sulfhydryl proximities to each other are within the
average range for disulfide bonding (alpha carbon = 3.0 - 7.5
angstroms) (Gao, et al.).
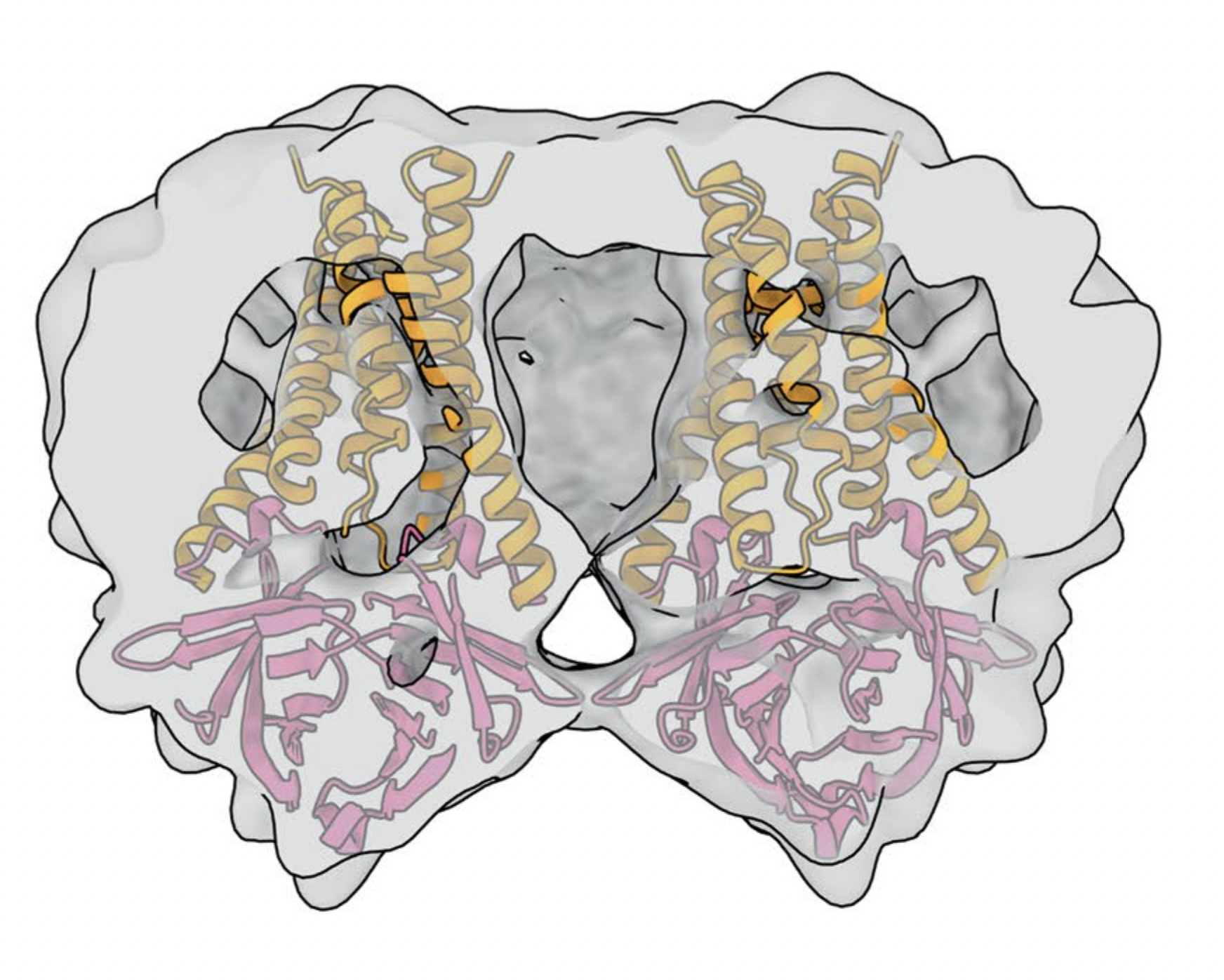
Figure 1. Macromolecule docking of an ORF3a tetramer, with a duplicate dimer positioned to the left of the JMOL-rendered ORF3a structure. (Source: Kern, et al.)
III. Ion Channel Activity
Biochemical assays reveal channels respond to potassium and calcium
ions, but deactivate in response to acidic pH conditions (Kern et
al.). Our rendered structure is most likely a "closed" state and would
undergo conformational changes to become a functional channel.
The inward facing side of the TM's are lined with residues that
create a polar cavity vertically through the center of the ORF3a
structure (Figure 2). There are also openings identified as "tunnels"
which connect the central polar cavity to the exterior of the protein,
shown in the figure below. The upper
tunnel (circumferenced by indicated
) leads into the intermembrane space; The lower
tunnel (circumferenced by indicated
) leads into cytosol.
Most ion channels also contain pores as a part of its ion conduction
pathway. These pores can resemble exterior divots or grooves
. For ORF3a, these are presented as hydrophilic regions formed in
between TM's. One particular groove is situated between TM2 and TM3,
indicated by the
. In the tetramerized form, these grooves in the dimer
interface could potentially also create an interior cavity, as
depicted in Figure 3.
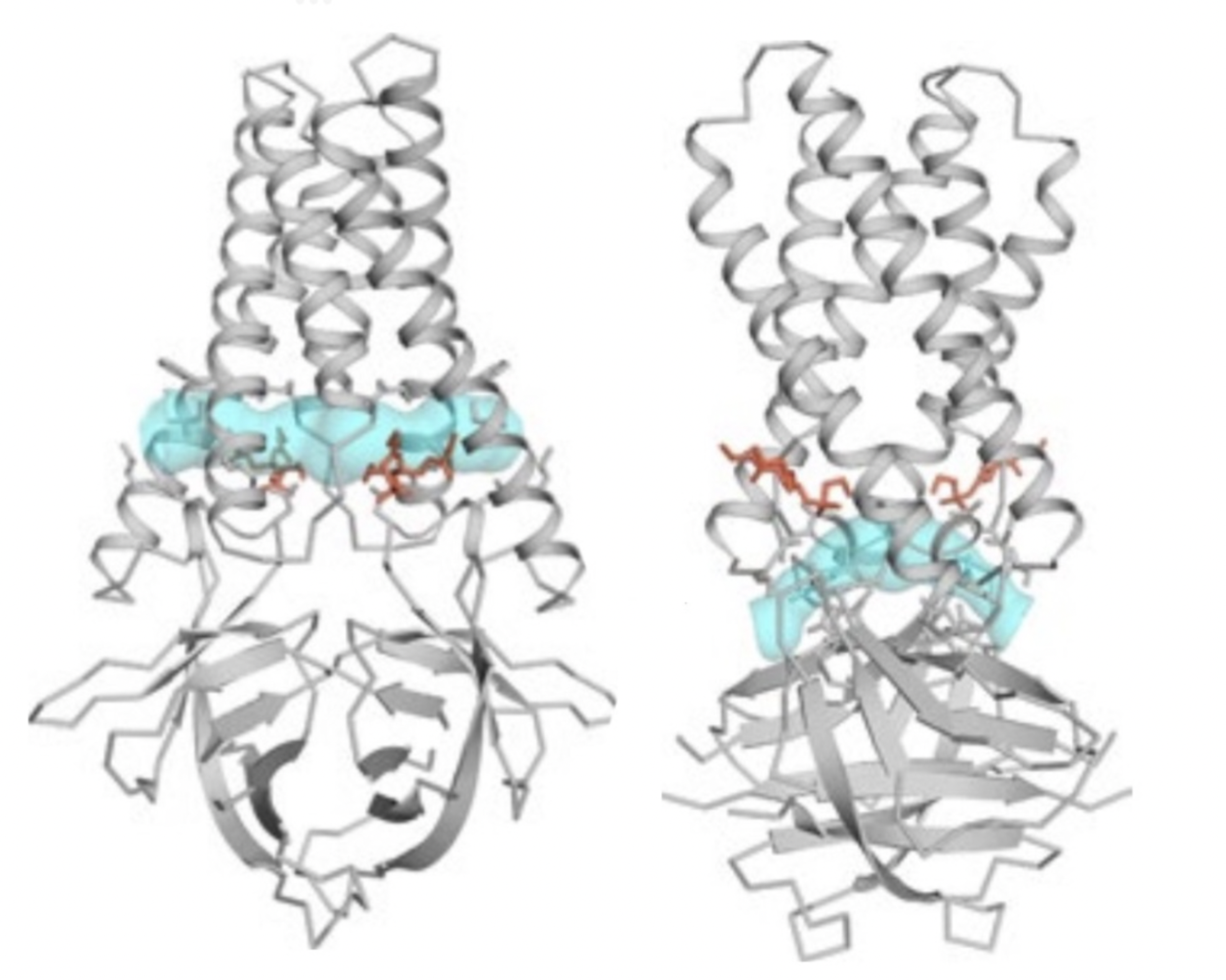
Figure 2. Digital rendering of upper tunnel (left)
and lower tunnel (right) in cyan blue in ORF3a structure. (Source:
Kern, et al.)
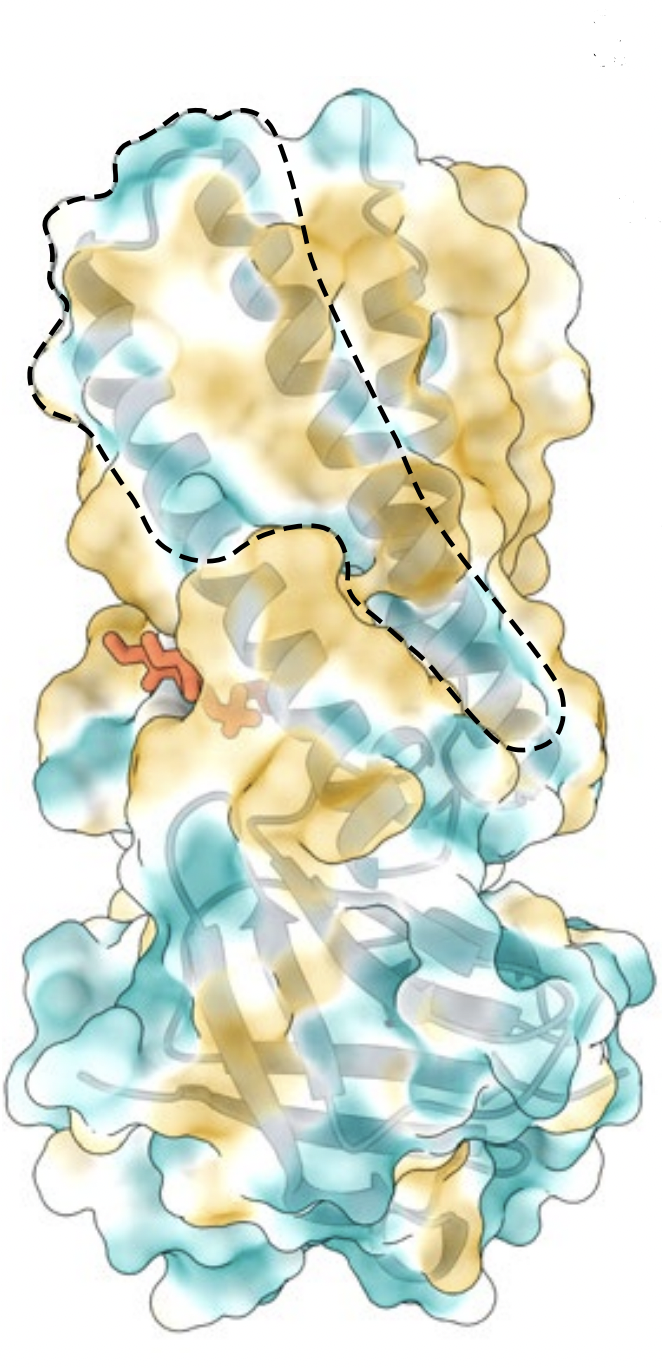
Figure 3. Solvent excluded surface rendering of
the TM2-3 hydrophilic groove (outlined in dashed lines). Color coding
demonstrates most hydrophilic regions (dark cyan) to most hydrophobic
regions (dark yellow). (Source: Kern, et al.)
IV. References
Breitinger, U., Farag, N. S., Sticht, H., and
Breitinger, H.-G. (2022). Viroporins: Structure, function, and their
role in the life cycle of SARS-CoV-2. The International Journal
of Biochemistry and Cell Biology, 145, 106185.
https://doi.org/10.1016/j.biocel.2022.106185
Gao, X., Dong, X., Li, X., Liu, Z., and Liu,
H. (2020). Prediction of disulfide bond engineering sites using a
machine learning method. Scientific Reports, 10, 10330.
https://doi.org/10.1038/s41598-020-67230-z
Kern, D. M., Sorum, B., Mali, S. S., Hoel, C.
M., Sridharan, S., Remis, J. P., Toso, D. B., Kotecha, A.,
Bautista, D. M., and Brohawn, S. G. (2021). Cryo-EM structure of
SARS-CoV-2 ORF3a in lipid nanodiscs. Nature Structural and
Molecular Biology, 28(7), 573-582.
https://doi.org/10.1038/s41594-021-00619-0
Ren, Y., Shu, T., Wu, D., Mu, J., Wang, C.,
Huang, M., Han, Y., Zhang, X.-Y., Zhou, W., Qiu, Y., and Zhou, X.
(2020). The ORF3a protein of SARS-CoV-2 induces apoptosis in
cells. Cellular and Molecular Immunology, 17(8), Article
8. https://doi.org/10.1038/s41423-020-0485-9
Back to Top