Biology Dept Kenyon College |
DNA Structure and Replication |

|
Biology Dept Kenyon College |
DNA Structure and Replication |

|
DNA Structure Note: Much of this modified from the MIT Hypertextbook. Deoxyribonucleic acid (DNA) was first identified in 1868 by Friedrich Miescher, a Swiss biologist, in the nuclei of pus cells obtained from discarded surgical bandages. The substance he found contained an acidic part, nucleic acid, and a basic (alkaline) part, which we now know to be histone proteins, which bind to the nucleic acid. Which component was the genetic material? Many scientists were sure that it was protein. After all, protein had so many subunits (20 amino acids) that it seemed obvious that there existed within protein the possibility for much more diversity in expressing the genetic code than in DNA, which only has 4 subunits. Each subunit is identical except for the base: Learn these structures. Click here for Quiz! DNA-binding proteins. In the long run, however, it turned out that the proteins attached to DNA have vital roles in chromosome structure and function. The roles of proteins include gene regulation (in bacteria and in eukaryotes) and chromosome imprinting.  Catabolite activator protein (blue and green) bound to promoter of lac operon. BIOL 363 Molecular Biology and Genomics
The Transforming Principle - DNA Might be the Genetic Material In 1943, Oswald Avery, Colin Macleod, and Maclyn McCarty, at the Rockefeller Institute, discovered that different strains of the bacterium Strepotococcus pneumonae could have different effects on a mouse. One virulent strain could kill an injected mouse, and another avirulent strain had no effect. When the virulent strain was heat-killed and injected into mice, there was no effect. But when a heat-killed virulent strain was coinjected with the avirulent strain, the mice died. What transforming principle was the dead virulent strain giving to the avirulent strain to make it lethal? This
phenomenon of
transformation,
the
uptake of DNA and incorporation into a genome, is now commonly
performed
in biotechnology..
Chargaff - Nucleotide Content in DNA In 1950, Erwin Chargaff at Columbia University discovered that no matter what tissue from an animal he looked at, the percentage content of each of the four nucleotides was the same, though the percentages could vary from species to species. In all animals:
%G = %C
The
significance of
these results was overlooked for three years, but they were crucial to
elucidating the structure of DNA.
Watson and Crick - The Double Helix In late 1953, James Watson and Francis Crick presented a model of the structure of DNA (see their paper in Nature.) It was already known from chemical studies that DNA was a polymer of nucleotide (sugar, base and phosphate) units. X-ray crytallographic data obtained by Rosalind Franklin, combined with the previous results from Chargaff and the chemists, were fitted together by Watson and Crick, who "borrowed" the data from Franklin's grant proposal. After several false starts, including the wrong tautomeric forms of the bases, they devised this model:
Under most cellular conditions, this double-stranded DNA molecule will coil naturally into a B-form helix, with one turn per 10.4 base pairs. However other structures are possible (see below). Each strand of the DNA is composed of nucleotides: The nucleotides form base pairs: 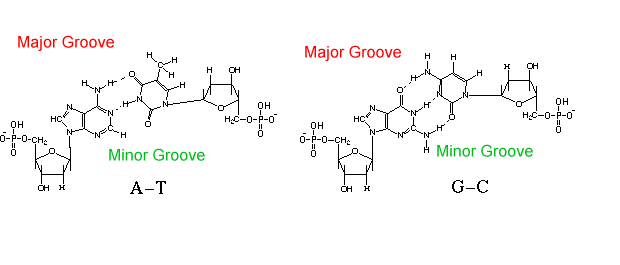 MIT Hypertextbook. Adenine
pairs with Thymine because they
make
two
hydrogen bonds.
The stacked base pairs form a major groove and a minor groove. Different regulatory proteins will bind to the major or minor groove. See Space-filling Model. Each base attaches to a phosphate at its 3' OH, and its 5'OH. The 2' carbon position has no OH; hence the "deoxy" part of DNA. The lack of 2' OH greatly stabilizes DNA, compared to RNA, because it prevents the intramolecular hydrolysis of phosphate linkages. The base
pairs "stack"
together like rungs on a ladder, because of favorable interactions
between
the pi orbitals extending out of the heteroaromatic ring structure of
each
base.
Helical forms of DNA The
structure of B-form
DNA helix was first determined by X-ray analysis of crystalized
molecules.
Certain regulatory sites within cells appear to have DNA sequence that takes a non-standard form, sometimes assisted by a protein. Moreover, DNA technologists are making use of unusual DNA properties to construct genetic medicines. Genetic medicines are pieces of artificial DNA that can hybridize to a region of the genome and turn off transcription of genes, such as a cancer gene. Stability
of DNA
However, in
water solution
certain chemical conditions can destabilize DNA.
Supercoiling In nearly all living cells, DNA contains negative superturns. This means it is "underwound," like a piece of yarn that has been twisted in the opposite direction that the multiple strands are wound. This is called negative supercoiling. Negative supercoiling may assist replication and transcription of DNA by lowering the energy needed to melt the helix. See the molecule Topoisomerase. In bacteria, negative superturns are maintained by the closed circular structure of the chromosome: It is impossible to unwind the superturns. In eukaryotes, negative superturns are maintained by the winding of the DNA helix around histone proteins. Problems. 1. Early life simulation experiments show that the base adenine would have formed spontaneously out of hydrogen cyanide, on the anaerobic early Earth. Show how five molecules of HCN can fit together to form exactly one molecule of adenine. 2. What kind of charge is on most of the proteins that take up 60% of the chromosome? Why? 3. If chemical analysis of a genome reveals 23% guanine, what are the percentages of the other three bases-- A, T, and C? 4. If a certain DNA site needs to come apart easily, for regulatory functions, what kind of base pairs are likely to be favored at that site? 5. Suppose an aromatic molecule with lots of pi orbitals could insert between two base pairs like a sandwich. What would happen as enzymes "read" the DNA information? 6. Some Archaea (microbes of the third Kingdom of organisms) living at extremely high temperature and pressure have positively supercoiled DNA. Why? DNA Replication DNA replicates semiconservatively. Replication starts by opening of the DNA helix at a particular sequence called an origin of replication (ori). Bacteria, even during logarithmic growth, have ONE map position where DNA can originate replication. Eukaryotes have MANY origins of replication, all of which run concurrently. In either case, each origin of replication runs bidirectionally, with TWO replicating forks. 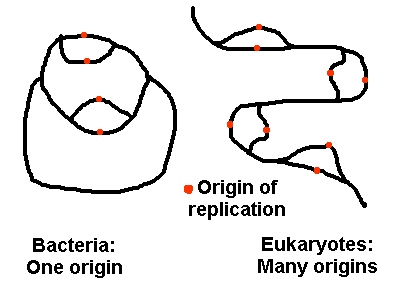 Bidirectional semiconservative replication can be demonstrated by observing DNA from cells replicating in the presence of radiolabeled nucleotides. Both sides of the dividing DNA will be labeled. What would the above diagrams look like, if the replicating DNA were radiolabeled? Molecular Steps of DNA Replication Replication of DNA is mediated by enzymes and binding proteins. One crucial function is to unwind the helix, enabling it to "unzip" exposing bases to pair with the growing strand. How can the helix be unwound without falling apart? To see an example, view topoisomerase I. DNA replication must be fast and accurate. To follow the step by step process, click the image: 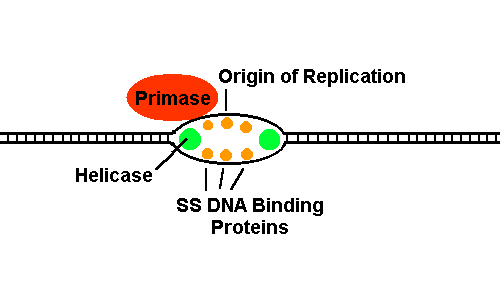 The helicase enzyme unwinds DNA, at the origin of replication. Two replicating forks are created. This reaction needs ATP. The exposed single-stranded DNA is protected by single-strand binding proteins (ssb). Click image for further steps. Leading and Lagging strands are replicated together
|