Biology Dept Kenyon College |
Gene Product Interaction, Crossover, Mobile Genes |

|
Biology Dept Kenyon College |
Gene Product Interaction, Crossover, Mobile Genes |

|
In real
living organisms,
all traits result from the interaction of many gene
products.
Suppose two enzymes are required to produce a trait. What may be
the result? What will happen to Mendelian inheritance ratios?
1. In sweet peas, purple is dominant; white is recessive. Suppose two recessive white flowers from different breeders are cross-bred together. The flowers--surprise--show the dominant purple color. When self-crossed, the ratio is 9 purple: 7 white. Explain these results. 2.
In fancy fowl, feathers are dominant to non-feathered. Two
pure-breeding
feathered birds from two different fowl fanciers are bred, producing
lovely
feathered offspring. The offspring are intercrossed, and a few of
their offspring have no feathers. The ratio is:
3.
Beetle colors. Suppose the color of beetles is determined by this enzyme pathway. Enzymes A and B are needed in series, but enzyme C represses enzyme A.  Figure out
what dihybrid
crosses would produce what ratios. Remember that you have to
specify
the entire genotype, including the purebreeding locus; for instance:
Complementation Complementation refers to the fact that mutations producing null alleles in different genes can be complemented, or compensated in effect, by the gene product provided elsewhere. An example of complementation is two-way recessive epistasis: The two white strains of sweet pea complement each other: P1 P1 p2 p2 X p1 p1 P2 P2 --> P1 p1 P2 p2 (purple flowers) You can use complementation analysis to intercross a large number of independently isolated null phenotypes, and figure out how many different genes are required for the enzyme pathway to produce the phenotype. See
the MIT site on Complementation.
Some forms of inheritance complicate the pattern of Mendelian inheritance and reassortment of traits. Inheritance may be overlaid by expansion of triplet repeats, or by parental imprinting. A few genes are not inherited by Mendelian principles at all, because they are contained on an extranuclear chromosome of the mitochondria or chloroplast. A well-known example is Fragile
X Syndrome. This very common cause of mental retardation
results from a region of repetitive DNA sequence that grows longer than
normal with each generation. The length of the sequence--a
multiple
triplet repeat, called a microsatellite--encourages
the region of the chromosome to shut down expression of an essential
FMR1
gene nearby. It also makes the chromosome "fragile;" that region
of the chromosome fails to condense during cell culture for
karyotyping.
Fragile X shows incomplete dominance; females who carry the trait will
show partial (but not total) compensation by the wild-type allele on
the
other X chromosome.
Imprinting and DNA methylation So far, all the inheritance mechanisms we have considered depend on DNA sequence of bases: A, T, C, G. However, certain sequences in addition contain chemical modification such as methylation. These modifications are added after DNA replication. They add information about how a gene will be expressed. In
mammals,
methylation occurs in the gonads, as sperm or eggs develop.
For example Huntington's disease is autosomal dominant. The onset of disease may occur in childhood, if you inherit the trait through your father; but only much later in life, if you inherit through your mother. Another disease involving methylation is Fragile X Syndrome. (See your handout.) In Fragile X, a region of repetitive CGG sequence is too long. The length of the repetitive sequence stimulates enzymes to "turn off" expression, including a nearby essential gene, FMR-1. This sequence is "turned off" through methylation in the female, and fails to get "turned back on" in her children. Children of males do not show the syndrome; but the grandchildren have high risk. Outside the nucleus are the mitochondria, which evolved out of endosymbiotic bacteria. Plants in addition havechloroplasts which arose the same way. Mitochondria have small circular chromosomes which are inherited through the female, because the sperm contribute no mitochondria to the fertilized egg. A
mother passes
on mitochondrial traits to all of her
children.
Some Mitochondrial
Diseases have been identified,
including
muscle degeneration and some forms of migraine headache.
Recombination by Crossover of Chromosomes Two DNA helices may "cross over" or recombine, a process requiring breakage of each strand at the phosphodiester backbone and ligation to another strand. There are two different classes of recombination:
Problem: Is hin recombinase involved in homologous recombination, or site-specific recombination? Explain your answer. Homologous recombination This image series shows how two homologous chromosomes within a tetrad cross over and exchange portions of their arms. (Problem: What phase of meiosis?) The blue and red arms designate homologues from different parents. (Why are they each double already?)Click the image: 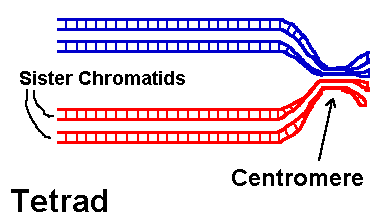
Linkage and Mapping What happens to Mendelian
ratios
when two genes are linked on the
same
chromosome?
A B
a
b
A
b
a B
A B
A
b
Test
Cross
45% A
B
5%
5% A
b
45%
5% a
B 45%
Which classes are "parental"? "Recombinant"? Map
Distances.
Problem: What happens when there are double crossovers? How does this interfere with map calculations? To practice gene mapping, try out Virtual Fly Lab. Problem: Try crossing female Stubble, Aristapedia with male wild type. Explain your results. (Note: both Stubble and Aristapedia traits have to start out hybrid, because the double dominant is lethal.) LOD scores and pedigrees. Today we use statistical analysis of human pedigrees to calculate the linkage between particular diseases and regions of DNA that may contain the gene for the disease. The "log of the odds" (LOD) score yields the probability of a given recombination frequence for a given set of pedigree data. Mobile genes. Some genes, such those encoding resistance to antibiotics, can move from one genome to another, at a new place in the genetic map. Some of these mobile genes can even transfer between two distantly related species of organism. Transposable
elements
A gene encoding antibiotic resistance. This gene confers a selective advantage to bacteria containing the transposon, in the presence of the antibiotic. Other transposed pieces of DNA can be inverted at one place in one species, to turn on or off the regulation of a gene. An example of such a site-specific transposition event is the flagellar gene regulation catalyzed by hin recombinase. Bacterial gene exchange differs from eukaryotes:
Transformation is the uptake of DNA from outside the cell. Only a single strand is taken up, through a special protein complex in the cell membrane. The process requires calcium ion (Ca2+). Transformation occurs at extremely low frequency, but with large populations of bacteria, it offers a significant route for genetic transfer. Phage
Transduction
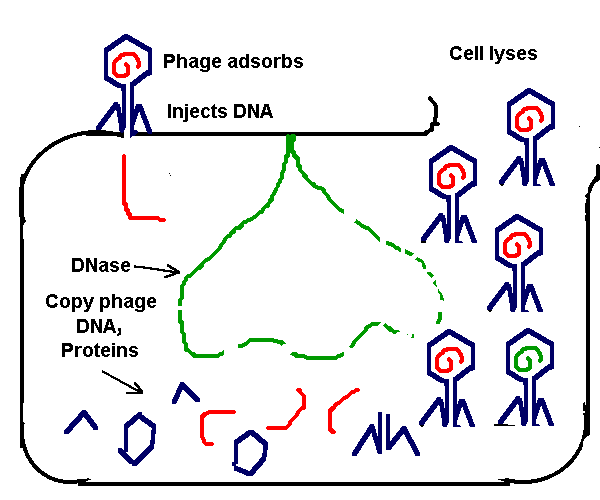
Plasmids are small circles of DNA that contain an origin of replication (ori) and a small number of genes, some of which may confer a survival advantage on a host. Some plasmids can transfer between different species; even between different kingdoms. A shuttle vector is a plasmid engineered in the test tube to contain an ori site for bacteria, and an ori site for animal or plant cells. Shuttle vectors are enormously useful to clone a gene conveniently in bacteria, then express it in tissue culture. Conjugation Conjugation is the process by which a plasmid is transferred from an F+ cell into an F- cell. The F factor in the F+ cell contains genes which express pili for attachment, and special membrane proteins for the transfer complex. Some conjugative plasmids carry drug resistant strains--a big problem for hospitals. If an F plasmid is integrated into a host genome (an Hfr, for high frequency recombination) the F factor can transfer part or all of the genome into the recipient F- cell. 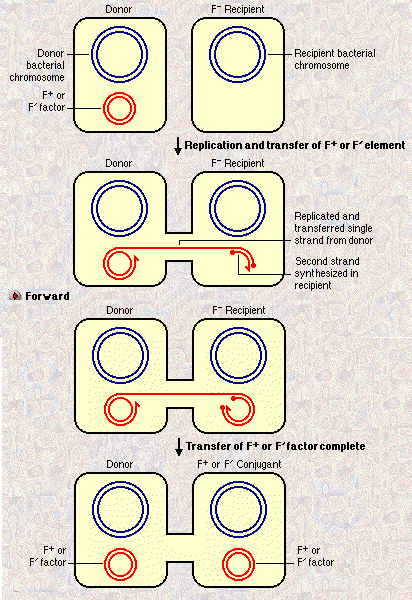 I I
Electronic Companion to Genetics, Cogito Learning MediaI The F plasmid can recombine itself into the host chromosome by site-specific recombination. It can then (a) transfer part or all of the chromosome into a recipient F- cell, as an Hfr; or (b) recombine itself out again, and mistakenly pick up a piece of the host chromosome to carry into the next F- host. Problem (5) Explain two different genetic processes in bacteria that can create a "partial diploid" for a small part of the genome. Explain why these processes are useful for bacterial genetic analysis. Interesting
sites on
bacterial genetics:
|