Bacterial Gene Transfer
Bacterial
gene exchange differs from eukaryotes:
- Bacteria
do not exchange genes by meiosis. (Why not?) They rarely
exchange two entire genomes.
- Bacteria
commonly exchange small pieces of genome, a few genes at a time,
through transformation, transduction, or conjugation.
- Transfer
between species, even kingdoms, is common; less common in
eukaryotes, though it does occur.
Transformation.
Transformation is the uptake
of DNA from outside the cell. Only a single strand is taken up,
through a special protein complex in the cell membrane. The
process requires calcium ion (Ca2+). Transformation occurs at
extremely low frequency, but with large populations of bacteria, it
offers a significant route for genetic transfer.
Phage Transduction
There are two types:
- Generalized transduction (depicted below). A piece of host DNA gets
packaged by mistake, instead of the phage DNA. This rare event
results in a phage delivering only bacterial DNA to the next
host. The DNA then recombines homologously, replacing the host allele.
- Specialized transduction, in which a lysogenic prophage recombines itself out
of the genome (by site-specific
recombination) and mistakenly includes
a piece of bacterial DNA. The resulting phage progeny can infect
cells to produce lysogens with a second
copy of the allele they had packaged, attached to the phage DNA.
Diagram of generalized transduction: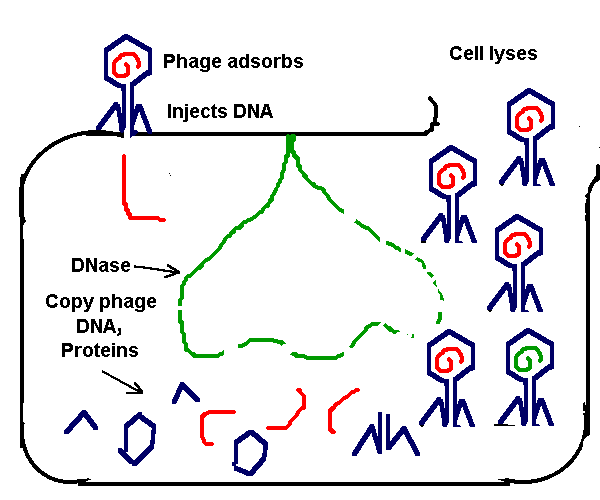
Plasmids
Plasmids are small
circles of DNA that contain an origin of replication (ori) and a small
number of genes, some of which may confer a survival advantage on a
host. Some plasmids can transfer between different species; even
between different kingdoms. A shuttle vector is a
plasmid engineered in the test tube to contain an ori site for
bacteria, and an ori site for animal or plant cells. Shuttle
vectors are enormously useful to clone a gene conveniently in bacteria,
then express it in tissue culture.
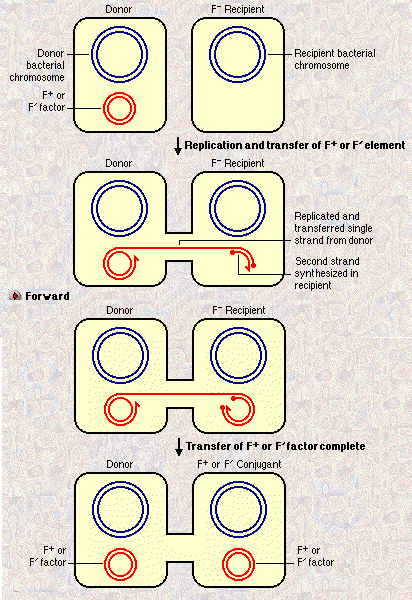
Conjugation
Conjugation is the process by which a
plasmid is transferred from an F+ cell into an F- cell. The F
factor in the F+ cell contains genes which express pili for attachment,
and special membrane proteins for the transfer complex. Some
conjugative plasmids carry drug resistant strains--a big problem for
hospitals.
If an F
plasmid is integrated into a host genome (an Hfr, for high frequency recombination) the F factor can transfer part
or all of the genome into the recipient F- cell.
 I I
Electronic Companion to Genetics, Cogito Learning MediaI
Episomes and Hfr
The F plasmid can recombine itself into the
host chromosome by site-specific recombination. It can then (a)
transfer part or all of the chromosome into a recipient F- cell, as an
Hfr; or (b) recombine itself out again, and mistakenly pick up a piece
of the host chromosome to carry into the next F- host.
Problem (5) Explain two different genetic
processes in bacteria that can create a "partial diploid" for a small
part of the genome. Explain why these processes are useful for
bacterial genetic analysis.
Mobile genes.
Some genes, such those encoding resistance to
antibiotics, can move from one genome to another, at a new place in the
genetic map. Some of these mobile genes can even transfer between
two distantly related species of organism.
Transposable elements
The first transposable elements to
be characterized genetically were controling elements for seed coat
color in maize (corn.) Barbara
McClintock won the Nobel Prize for showing that DNA is not all
"fixed" in the genome, but that some sequences can insert and excise by
intramolecular recombination.
There
are many classes of transposons.
In bacteria, a common structure of a transposon contains:
- An insertion sequence (IS) at the right and left ends. The IS contains the
gene encoding the transposase enzyme.
- A gene encoding antibiotic
resistance. This gene confers a selective advantage to bacteria
containing the transposon, in the presence of the antibiotic.
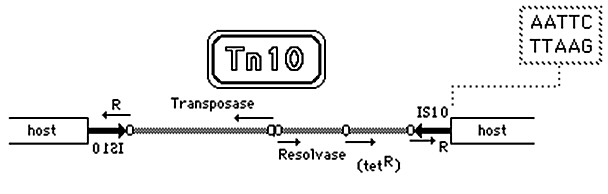
Transposons
Some bacterial transposons can be exchanged among many different species, usually carried by plasmids.
Other transposed pieces of DNA can be inverted at one place in one
species, to turn on or off the regulation of a gene. An example
of such a site-specific transposition event is the flagellar gene
regulation catalyzed by hin
recombinase.
Genomic Islands
| Bacterial genomes often contain "islands" of DNA transferred
relatively recently from another species. The "genomic
island" may confer special properties to a pathogen, or two a
strain inhabiting a special niche in an ecosystem. |
Herbert Schmidt and Michael Hensel, Clinical
Microbiology Reviews, January 2004, Vol. 17, p. 14-56.
|
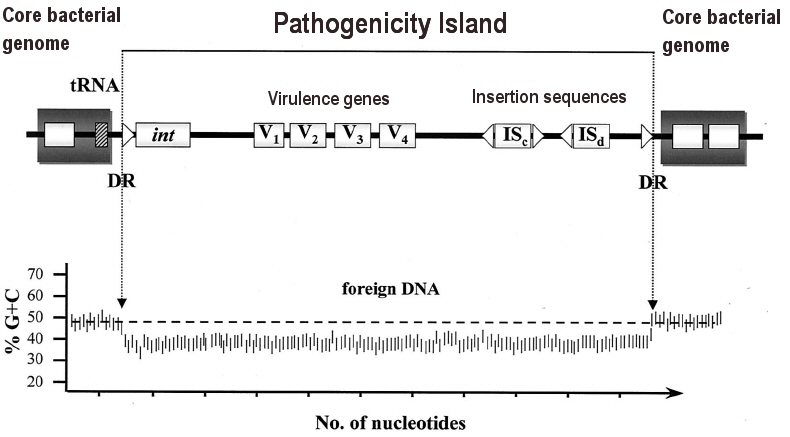
A pathogenicity island is a genomic island that
converts a "harmless" bacterium to a pathogen.
The
pathogenicity island is a distinct region of DNA present in the genome
of pathogenic bacteria but absent in nonpathogenic strains of the same
species. Transfer may be mediated by an integrase
enzyme (int).
The
island is typically inserted at a tRNA gene
in the core genome.
The
pathogenicity island typically contains virulence
genes (V1 to V4)
interspersed with other mobility elements, such as insertion sequences (IS).
How do we recognize a recently inserted pathogenicity island?
One clue typically is a difference in percent GC
content, compared to the core genome.
|
Examples of
Pathogenicity Islands
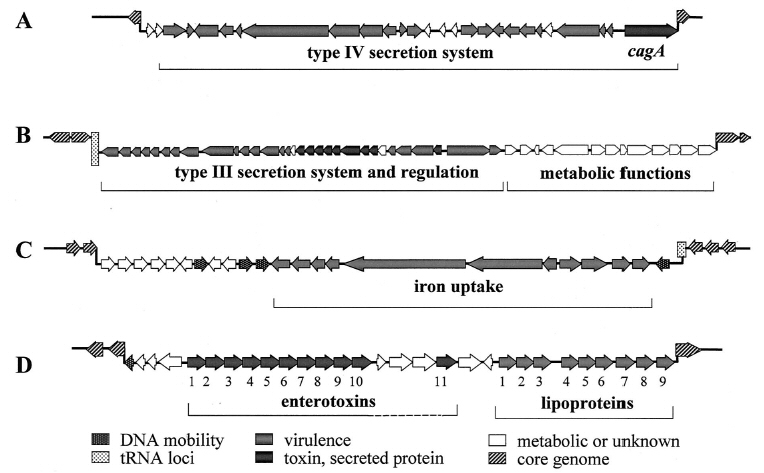
|
A.
The cag island of H. pylori
(cause of stomach ulcers) harbors genes for a type IV secretion system
that can translocate the toxin CagA into human cells, causing an
inflammatory response.
B.
The SP-1 island of Salmonella typhimurium (typhus and food
poisoning) encodes a type III secretion system (grey), secreted
proteins (dark grey), and regulatory proteins. The same island includes
metabolic proteins unrelated to virulence.
C.
The HPI island of Yersinia enterocolitica has genes that
encode a high-affinity iron uptake system (dark grey) needed for
extracellular growth of the pathogen during colonization of the host.
D.
The vSAL island of multiple drug-resistant Staphylococcus
aureus (MRSA) encodes a remarkably high number of enterotoxins.
|
Genomic islands in two
strains of a marine phototroph, Prochlorococcus
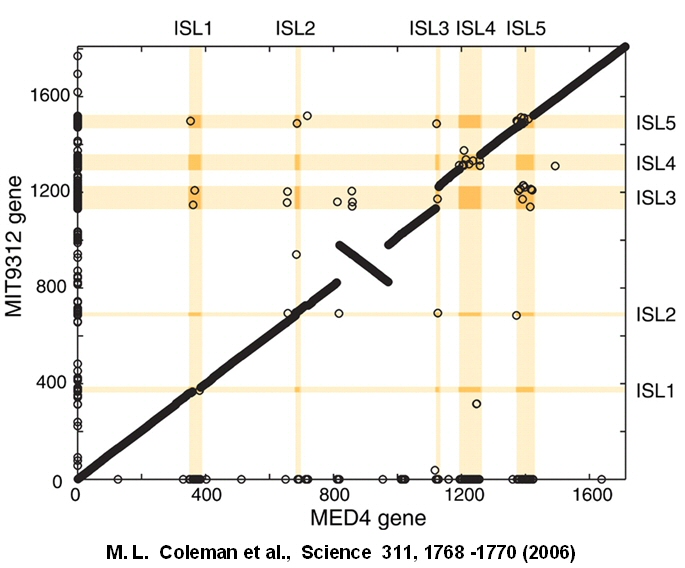
The
two strains of Prochlorococcus marinus are cyanobacteria,
major oxygenic producers in the oceans, consuming a large part of
atmospheric CO2. The strains MED4 and MIT96512 differ by
only 0.8% of their genome, yet their distributions throughout the ocean
are very different, for unknown reasons. The reason for the difference
in distribution may have to do with genes encoded within five genomic
islands specific to MED4 (ISL1, ISL4, ISL5) or to MIT9312 (ISL2, ISL3).
|



 I
I