Biology Dept Kenyon College |
Genetic Code, Translation, Splicing |

|
Biology Dept Kenyon College |
Genetic Code, Translation, Splicing |

|
The Genetic Code How do 64 different codons produce 20
different amino
acids?
Evolution of the Code
Translation involves the conversion of a four base code (ATCG) into twenty different amino acids. A codon or triplet of bases specifies a given amino acid. Most amino acids are specified by more than one codon. The conversion of codon information into proteins is conducted by transfer RNA. Each transfer RNA (tRNA) has an anticodon which can base pair with a codon. Some anti-codons have modified bases that can pair with more than one codon, specifying the same amino acid; this means that we don't need 61 different tRNA molecules for all 61 codons. (What do the other three codons specify?) The structure of transfer RNA (tRNA): 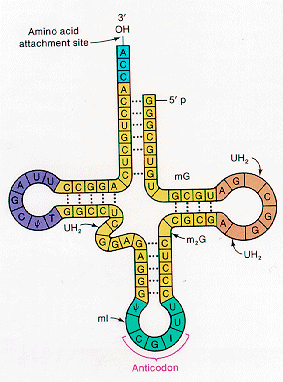 Transfer RNA (tRNA) has the following structure:
H2N--C--C--OH + HO--tRNA --> H2N--C--C--O--tRNA + H2O This process, called charging, is catalyzed by a tRNA transferase, or aminoacyl tRNA synthetase, specific to the tRNA type. There are one or more tRNA types, specified by different genes, for EACH amino acid.
Translation of mRNA into polypeptide Translation requires initiation, elongation, and termination. Translation is performed by the ribosome, an organelle composed of more than fifty different proteins plus two structural rRNAs, each part of the 30s subunit or the 70s subunit. The "s" is a unit of sedimentation, referring to how fast a particle settles out during centrifugation. Note that this entire process requires tRNAs continually being charged with their respective amino acids, by tRNA transferase enzymes. (1) Initiation occurs by binding of the 30s subunit to the mRNA. In bacteria, the mRNA binds by hybridization of a special sequence to the Shine-Dalgarno sequence of the 16s rRNA, part of the 30s subunit. The ribosome then finds the first AUG sequence on the mRNA, where it binds the anti-codon of a Met-tRNA, at the P site. (2) Elongation occurs by successive amidation of the nascent (growing) chain. The 50s subunit now binds, creating the A site. Each new aminoacyl-tRNA enters at the A site, where it transfers the amino end of its amino acid to the carboxylic end of the nascent chain. The entire ribosome now "translates" over one codon position, so that the nascent chain is now bound to the P site. Elongation requires energy provided by GTP. (3) Termination occurs when the A site reaches a stop codon. Since no tRNA exists with an anticodon complementary to the stop codon, the ribosome "pauses" until at last it "falls off" the mRNA, and the polypeptide chain terminates. This process is facilitated by a release factor protein that binds into the ribosomal A site containing a stop codon to help with protein release. 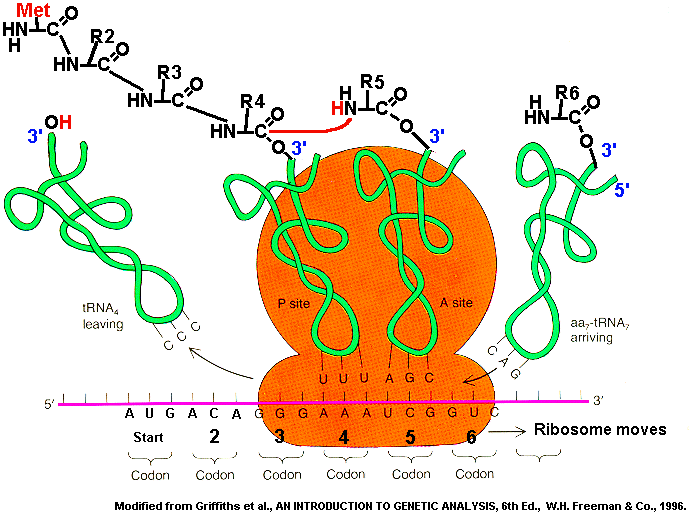
Where does the mRNA come from? As soon as mRNA starts getting transcribed, ribosomes attach to translate:
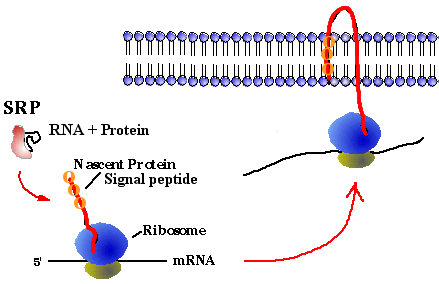
Where does the growing peptide go? If the growing peptide is water-soluble, to function in the cytoplasm, it folds itself into its native comformation, with the help of chaperone proteins. If the growing peptide is hydrophobic, to function in the membrane, its hydrophobic signal peptide attaches to the signal recognition particle (SRP). The SRP is composed of protein and RNA (like the ribosome). SRP carries the hydrophobic peptide, with its ribosome, to the face of a membrane for insertion:
Online practice problems: Try problems 9 - 15 of the Nucleic Acids Problem Set from the Biology Project at the University of Arizona. Energy considerations Problem: DNA replication, RNA
transcription,
and protein translation take lots of energy. Why?
Animation: Translation from U. Conn. In eukaryotes, production of mRNA is more complicated than in bacteria, because:
A complication in eukaryotic transcription is the existence of three different RNA polymerases, which transcribe three different classes of genes. RNA pol II transcribes hnRNA (precursor to mRNA). RNA pol I and III transcribe functional RNAs such as rRNAs and tRNAs. Splicing of hnRNA to make mRNA
The splicing of introns is a complex intramolecular reaction, mediated by an organelle composed of RNA and protein molecules, the spliceosome.The spliceosome catalyzes the reaction between a 2'OH of an Adenine, and the 5' phosphate end of the intron, creating a lariat loop. (Note: Only RNA has 2'OH to do this!) The lariat reaction produced a 3'OH on one exon, enabling to join the 5' phosphate of the joining exon. The spliceosome is actually composed of several small nuclear riboprotein (RNA-protein) organelles, called snRNPs. To see how these snRNPs (labeled U1, U2 etc.) splice out the intron, view this animation. Click on each image. Why would cells have evolved to have introns, which seem wasteful of DNA an energy? Two kinds of reasons have been proposed:
Alternative Splicing Alternative splicing of cardiac troponin T (cTNT) gene during development
|