Electrophoretic
Analysis of DNA **Practice Plasmid.exe -- It will be on the
Quiz**
Cloning
genes
PCR
Gene
sequence analysis
Complementarity
and Hybridization
Northern,
Southern, and Western blots
DNA
Microarrays
Transgenics
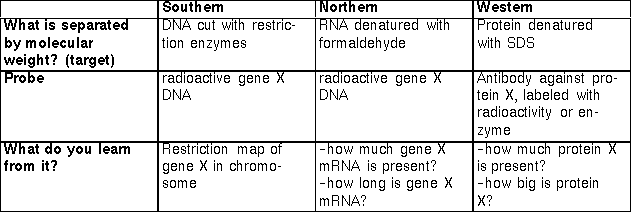
The molecules to separate (DNA RNA) carry a net negative charge (why?) so they move along the electric field toward the positive cathode. (To separate proteins, a detergent would be included which coats the protein with negative charge.)
The larger molecules are held up as they try to pass through the pores of the gel, while the smaller molecules are impeded less and move faster. This results in separation by size, with the larger molecules nearer the well and the smaller molecules farther away.
Note that this separates on the basis of size (volume in solution), which is not necessarily molecular weight. For example:
Sample 1 contains only one size class of macromolecule - it could be a plasmid, a pure mRNA transcript, or a purified protein. In this case, you would not have to use a probe to detect the molecule of interest since there is only one type of molecule present. Blotting is usually necessary for samples that are not complex mixtures. By interpolation, its molecular weight is roughly 3.
Sample 2 is what a sample of total DNA cut with a restriction enzyme, total cellular RNA, or total cellular protein would look like in a gel stained with a sequence-independent stain. There are so many bands that it is impossible to find the one we are interested in. Without a probe (which acts like a sequence-dependent stain) we cannot get very much information from a sample like this.
Different stains
are used for different classes of macromolecules. DNA and RNA are
generally stained with ethidium bromide (EtBr),
an intercalating agent. The DNA-EtBr complex fluoresces
under UV light. Protein is stained with Coomassie Blue or
Silver Stain.
Since then, the dangers have appeared to be little more than those of "natural" genetic mixing. But we remain concerned about issues such as:
One approach would be to cut the appropriate gene from human DNA and paste, or splice, it into a vector such as a plasmid or phage DNA. Our "scissors" are the class of enzymes called restriction endonucleases
Restriction
Endonucleases
An "endonuclease" is an enzyme that
cuts duplex DNA in the middle, not at an end (for
exonuclease). Different species of bacteria have evolved
different restriction endonucleases, each to cut foreign DNA that gets
into their cells by mistake. To be cut, the DNA has to lack their
own pattern of protective methylation. There are well over
a hundred restriction enzymes, each cutting in a very precise way a
specific base sequence of the DNA molecule.
A restriction endonuclease cuts DNA only at a specific site, usually containing 4-6 base pairs. The enzyme has to cut the DNA backbone twice, recognizing the same type of site; therefore, the site "reads" the same way backwards as forwards--a palindrome.
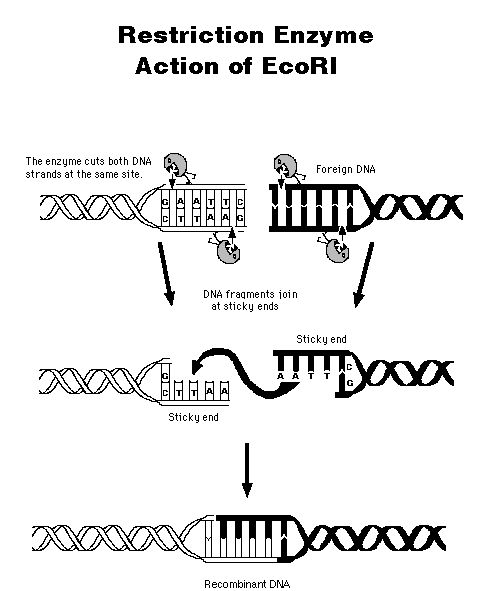
This "sticky ends" from two different DNA molecules can hybridize together; then the nicks are sealed using ligase. (Where does ligase come from? What is its natural function?) The result is recombinant DNA. When this recombinant vector is inserted into E. coli, the cell will be able to process the instructions to assemble the amino acids for insulin production. More importantly, the new instructions are passed along to the next generation of E. coli cells in the process known as gene cloning.
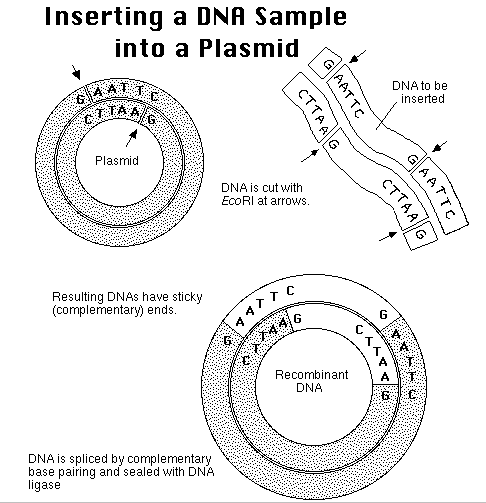
Restriction site
Analysis
How can we use restriction sites to
analyze the plasmid products of ligation, and tell whether we in fact
have ligated the correct molecule:
Problem: Suggest
several "incorrect" ways the plasmid could recombine.
More Problems:
Use the PLASMID Program. You MUST practice restriction
analysis with this program; it will be on the quiz and/or the test.
How do we get the recombinant molecule into a bacterial cell? Usually by transformation (for a plasmid) or by in vitro packaging into a phage head coat (for a phage vector such as lambda phage).
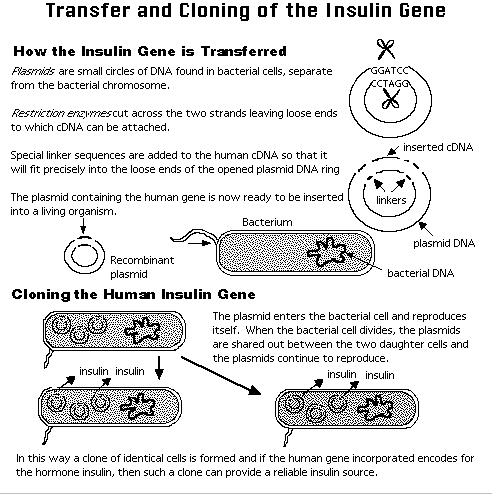
The above is a highly simplified
description of recombinant DNA technology.
How would we actually locate the
appropriately cloned gene? There are many different ways,
depending on the specific case. Here is one example, in which a
partial sequence of the protein enables us to reverse
the code and determine an approximate DNA
sequence to use for a radiolabeled
probe. The DNA probe will hybridize to clones
containing the correct DNA, even if it is just one piece cut out of an
entire genome.

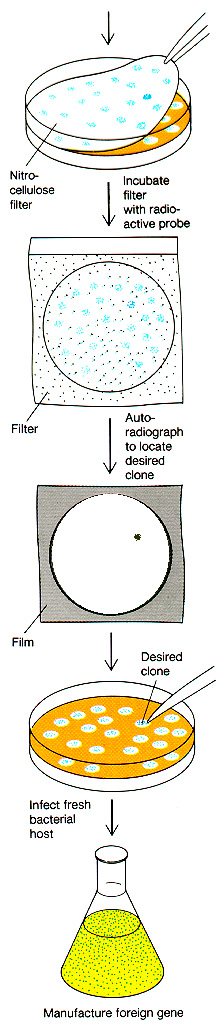
The radioactive probe is made by determining a short segment of the protein sequence, then "back translating" to the possible DNA sequences. Short DNA sequences are synthesized to match the protein sequence. Then these DNA oligomers (known as "oligos") are radiolabeled, and applied to the blotted clones. They should hybridize only to clones containing sequence encoding the desired protein.
Reverse
transcription: cDNA Cloning
Suppose we need to clone a gene
containing lots of introns. What will
happen when the bacterium tries to express it?
To overcome this
problem, we can start with mRNA isolated from tissues that produce the desired
protein. We then use reverse
transcriptase enzyme (produced by a
retrovirus related to HIV) to reverse transcribe the mRNA into a DNA
molecule that now is free of introns. Now we can ligate "sticky
ends" onto the cDNA and recombine it into a phage or plasmid vector.
Problem:
Know the differences between genomic cloning and cDNA cloning.
Explain the
relative ADVANTAGES and DISADVANTAGES of each technique--depending on
the aim of your research.
In PCR, a heat-stable DNA polymerase is used, most commonly Taq Polymerase from the thermophilic microbe Thermus aquaticus. Thomas Brock discovered T. aquaticus from a hot spring at Yellowstone National Park.
 Prismatic
Pool, Yellowstone
Prismatic
Pool, Yellowstone
More recently, an even more heat-resistant polymerase has been developed from a hyperthermophilic microbe growing at 110 degrees C in hydrothermal vent ecosystems in the deep ocean; it's called "Vent Polymerase."
The Taq Polymerase is put with the DNA to be amplified, plus all four NTPs, plus two primers facing each other, about 200 - 6000 kb apart. (Why do we need primers?) The primers are selected based on the DNA region you want to amplify. The tube is placed in a thermal cycler.
DNA gets synthesized from each primer, for about 2 minutes. Then the temperature is raised to 95 degrees C -- enough to denature (split apart) the DNA base pairs. But the Taq Polymerase remains intact, because it comes from an organism that evolved to grow at this temperature.
Now the temperature is decreased again, and primers again can hybridize to the DNA--both the old AND the newly synthesized strands. Again, Taq Polymerase extends new DNA strands. Again, the temperature is raised.
After repeated cycles, the amount of DNA sequence between the two primers increases exponentially. First 2 strands, then 4, 8, 16, up to about a million. Thus, in a couple of hours, you can get million-fold amplification of a DNA sequence.
Applications of PCR
PCR has replaced cloning for many
purposes, particularly the sequencing of DNA. It is faster and
requires no vectors, which can mutate as they reproduce. It
can be used forensically, to amplify tiny amounts of DNA from criminal
evidence; or clinically, to detect DNA sequences linked to inherited
disorders.
|
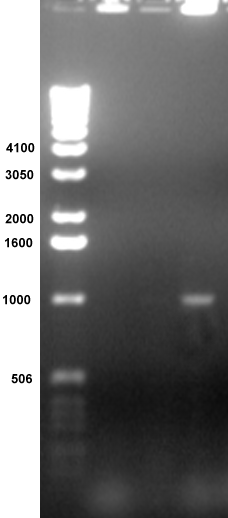
Microbiology Lab: Students characterized the microbial species of various environments in Knox County. Microbial colonies were placed directly into PCR reactions containing primers for amplification of ribosomal RNA genes (rDNA). Lane 4 contains an amplified band of the predicted size, 1000 bp. The top of each well contains genomic DNA. The smears at the bottom contain the PCR primers. The DNA will be sequenced and matched through GenBank to determine the microbial genus. Thanks to Dan Nickerson '00 and Adam Marks '01. |
 |
The main limitations of PCR are:
The sequence of DNA base pairs can be analyzed by
Once we have a piece of DNA cloned, it
is amplified (available in many copies)
and we now have a living clone which
provides, in theory, an indefinite source of the DNA sequence.
How do we analyze the sequence?
DNA Sequence
Determination
Once you have identified a particular
region of DNA of interest, you need to find out the precise sequence of
DNA nucleotides. This is done by di-deoxy
sequencing, in which a DNA polymerase is put together with dNTPs
in four different reactions, each containing a small amount of one
di-deoxy NTP (ATP, TTP, CTP, or GTP). The di-deoxy nucleotide
lacks a 3'OH to continue chain extension, so the chain
terminates. Each reaction produces a population of fragments
terminated at A, T, C, or G.
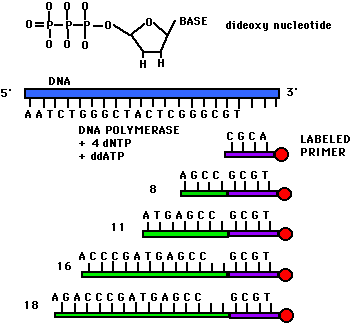
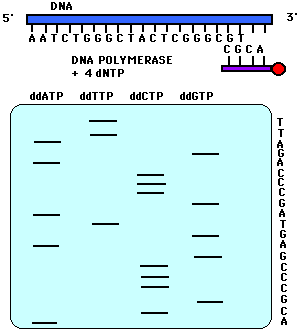
images from Davidson College
In solution, hybrid molecular complexes (usually called hybrids) of the following types can exist (other combinations are possible):
These molecules must then be immobilized on a solid support, so that they will remain in position during probing and washing. The probe is then added, the non-specifically bound probe is removed, and the probe is detected. The place where the probe is detected corresponds to the location of the immobilized target molecule. This process is diagrammed below:
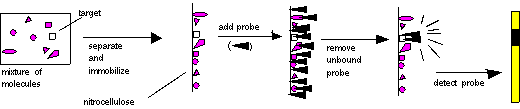
In the case of Southern, Northern, and Western blots, the initial separation of molecules is done on the basis of molecular weight, by gel electrophoresis.
We can now put most of the protein-encoding genes onto a microarray chip, using technology based on the DNA silicon chip industry. The chip can be used to hybridize to cellular RNA, and measure the expression rates of a large number of genes in a cell. More details here.
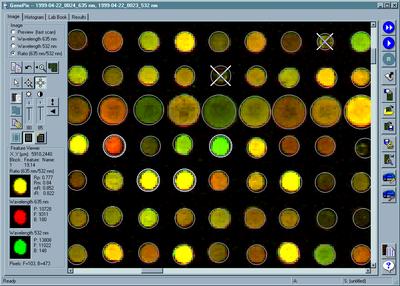
Axon Industries.
From "Everything's Great When It Sits on a Chip," The Scientist,
Volume 13, #11, May 24, 1999
Cloning in Animals.
Animals have two different classes of
cells: germ line and somatic.
Only alterations in the germ cells
can be transmitted to future generations. However, some forms of somatic cell gene therapy
can be useful in treating patients. For example, people with
cystic fibrosis can receive the cloned CFTR gene in a nasal spray,
which then infects the lining of their lungs and improves lung
function. The infected (transformed) epithelial cells eventually
are lost, however, during normal processes of tissue growth, so the
treatment needs to be repeated.
Germ-line cloning. There are two basic ways to clone a gene in the mammalian germ line.
But when you create a clone, how do you know it worked? You need to know:
>>Back to Syllabus>>