Hemagglutinin (HA) - Cell Binding protein
in Avian Influenza
Jack Cerchiara,'06 and Brendan Holsberry, 07
Contents:
I. Introduction
The Influenza A virus is an orthomyxovirus, and
its receptor binding complex is comprised of two primary structural proteins,
Hemagglutinin (HA) and Neuraminidase (NA). It has been determined that Hemagglutinin
is the primary protein responsible for binding to receptor sites on the
cell membrane, allowing the virion to enter the cell (Subbarao 2000). Hemagglutinin
is species specific binding protein that binds only to matched sialic acid
receptors in host cells (Subbarao 2000). The molecule under study here is
the HA protein extracted from the H1-human influenza strain.
The structure of Hemagglutinin is very similar between strains,
and differs in only a few structural difference. What makes this strain
differ from the H5N1 influenza now posing a pandemic threat is the speficity
of its binding regions, which will be the basis of this study. What makes
this strain of influenza so dangerous is both its rapid ability to evolve
and the fact that mammals, specifically humans contain to immune defenses
against this avian strain (Suarez 2000). In 1918, a strain of influenza
A virus killed 20-40million people worldwide and the World Health Organization
estimates nearly 7 million dead and 1 billion ill from an outbreak of the
current H5N1 strain (Horimoto 2001).
Why look at Hemagglutinin?
Researchers have been able to determine that Hemagglutinin (HA) is
a species specific binding protein that allows for the virus to bind to
the cell membrane of host respiratory cells and propagate through cellular
processes. By examining this process, medical researchers hope to determine
a vaccine that may prevent this binding from occurring, thus preventing
host infection.
II. General Structure
Hemagglutinin is a trimer protein composed of a globular
domain and a stem domain, divided along
the longitudinal axis of the protein
. As was stated, HA protein is made up of three monomers;
HA1, HA2,
and HA3 domains
. Each of these monomers is comprised
to two subdomains, in the stem domain the two helix are bonded at Phe-88
to Phe-63 a clear difference between
influenza strains, in the avian H5N1, it displays a inward facing Phenylalanine
ring, while H1- human strains display an outward facing ring shown here.
Along the longitudinal axis, the protein is comprised of structural alpha-helices
and beta sheets
are seen especially in forming the "bonding depression"
in the globular region which will be discussed in the following section.
Note the beta-sheets are primarily present
in the globular head
where the binding region to sialic acid resides and the alpha
helices
make up the stem region of the HA monomers .
III. Receptor Binding Sites
The hemagglutinin protein, as was previously stated, is
a trinomer protein, primarily responsible for the binding of the Influenza
A virion to cell surface receptors, membrane fusion and intracellular infection,
which is the first stage of viral infection. Hemaggluntin recognizes sialic
acid components of cell-surface glycoproteins and glycolipids (Gamblin et
al., 2004; Ha et al., 2000). Hemagglutinin contains a shallow
depression at its "head"
which allows the sialic acid sequence to move into like a "lock
and key." According to Ha et al. (2000), species
specific sailic acid receptor analogs do not have a binding affect on the
orientation of the sialic acid domain into the base of the hemagglutinin
depression. One side of the sialic acid's pyranose ring faces the base of
the binding depression. The axial carboxylate, acetamido nitrogen, and the
8- and 9-hydroxyl groups face the site and form bonds.
The binding depression surrounds the sialic acid domain with three
primary regions of the hemagglutinin structure. This region is comprised
of a loop-helix-loop structure, which surrounds
the sialic acid. The 130-loop, 190-helix
and 220-loop
structures form the triangular opening into the beta-sheet depression.
These three structures from the primary binding to the sialic
acid and specific regions in their domains (Gamblin et al., 2004).
The exact binding geometry differs between species specific hemagglutinin
and cell surface proteins, however there are primary cites on the loop-helix-loop
complexes that form specific integrations allowing for the binding of the
hemagglutinin and the subsequent cell infection. In human-avian receptor
complexes the Glu-190
residue on the 190-helix
forms a hydrogen bond to the 9-hydrxyl group.
Thr-136
as well as amino-acids at residues 135
and 137
on
the 130-loop form hydrogen bonds
to the sialic acid's carboxylate. Also, Lys-222
and Gln-226
of the 220-loop
form bonds with 8-hydroxyl group of the sialic
acid (Gamblin et al., 2004) (Figure 1). Together this binding forms
around the sialic acid domain of the cell surface glycoprotein or glycolipid
in the HA depression, connecting each monomer to the sialic acid on the
cell, initiating viral infection
.
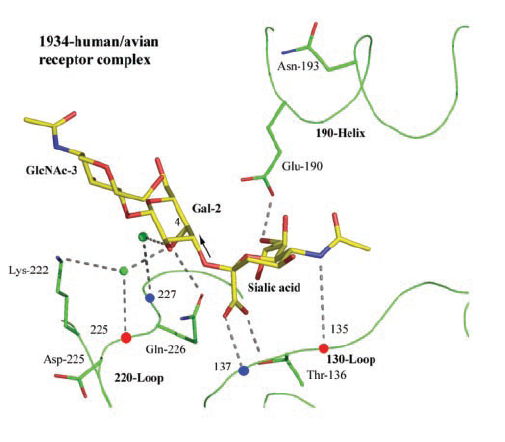
(Gamblin et al., 2004)
IV. Species Specific Binding
The HAs of viruses recognize different linkage if the HA strain is
avian or human. Avian strains have receptor–binding specificity for
sialic acid receptors in alpha-2,3 linkage while human strains have specificity
for alpha-2,6 linkage. Avian H5 HA hydrogen bonds through Gln-226 to the glycosidic
oxygen that is exposed in a trans coformation in the alpha ;2,3 sialic acid-to-galactose
link. Alpha-2,6-Linked sialosides bind in a cis conformation, exposing the
glycosidic oxygen to solution and nonpolar atoms of the receptor to Leu-226,
a human-specific residue. Although each strain contains the both types of
alpha linkages, the Gln-226/Gly-228 of the avian strain prefer the alpha-2,6
linkage while the Leu-226/Ser-228 of the human strain prefers the alpha-2,3
linkage. The two can geometrically distinguished because the human strain
is much wider between the 226 and 228 positions while the avian strain is
much more closed.
V. References
- Ya Ha, David J. Stevens, John J. Skehel, and Don C. Wiley.
X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins
bound to avian and human receptor analogs. PNAS, Sep 2001; 98: 11181.
- S. J. Gamblin, L. F. Haire, R. J. Russell, D. J. Stevens,
B. Xiao, Y. Ha, N. Vasisht, D. A. Steinhauer, R. S. Daniels, A. Elliot, D. C.
Wiley, and J. J. Skehel
The Structure and Receptor Binding Properties of the 1918 Influenza
Hemagglutinin. Science, Mar 2004; 303: 1838 - 1842.
- Sauter, N. K., Hanson, J. E., Glick,
G. D., Brown, J. H., Crowther, R. L., Park, S. J., Skehel, J. J., Wiley, D.
C.: Binding of influenza virus hemagglutinin to analogs of its cell-surface
receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy
and X-ray crystallography. Biochemistry 31 pp. 9609 (1992)
- Subbarao, K.; Katz, J. Avian influenza viruses infecting
humans. Cellular and Molecular Life Sciences. Vol: 57, Issue: 12, November,
2000, pp. 1770 - 1784
- Suarez, David L. Evolution of avian influenza viruses
Veterinary Microbiology, Vol: 74, Issue: 1-2, May 22, 2000 pp. 15-27
- Horimoto, Taisuke; Kawaoka, Yoshihiro. Pandemic
Threat Posed by Avian Influenza A Viruses. Clinical Microbiolical Review.,
Jan 2001; Vol. 14: pg. 129 - 149.