Human Salivary Amylase
Alexandra Simic
Hachmann '17
Daniel Maffezzoli '17
Contents:
I. Introduction
Model View:
Human
salivary alpha amylase (HSAmy) is an important enzyme found in the
oral cavity. It belongs to the glycoside
hydrolase family and exists in various isoforms in salivary
secretions. Humans produce two kinds of alpha amylase (salivary and
pancreatic amylase) that overall share about 97% homology.
HSAmy is a type of hydrolase that breaks down complex
carbohydrates into simple sugars, like glucose or maltose, by
cleaving alpha-1,4-glucosidic bonds. Further breakdown of the starch
is completed by pancreatic amylase later in digestion.
This enzyme has distinct components including a calcium ion,
chloride ion, and various amino acid residues that play critical
roles in hydrolytic activity and substrate binding.
HSAmy can also bind to oral streptococci
bacteria. This results in the bacteria either being removed from or
stored in the oral cavity. Storage and accumulation of the bacteria
in the oral cavity causes dental complications like plaque and tooth
decay.
II. General Structure
The structure of HSAmy consists of a single polypeptide
chain of 496 amino acids that can be divided into three domains.
houses the active site and contains three catalytic residues: Asp197,
Glu233,
and Asp300.
The neon structures are GLC sugars used for crystallography
purposes and demonstrate the binding region. Hydrophobic ligands Arg337,
Arg195 and
Asn298 function as binding sites for chloride ions,
which are
required for full catalytic activity. These binding sites also
contain hydrophobic residues (
Phe265
and Phe295) nearby
that aid in the catalytic activity of the enzyme. Only one
chloride ion and calcium ion bind per molecule of HSAmy.
consists of one calcium binding site. HSAmy is a calcium
metalloenzyme and therefore cannot function efficiently in the
absence of calcium. The
ion
serves as a stabilizer during hydrolytic activity and is
held in place by residues Arg158,
Asn100, Asp167,
and His201.
is arranged in a Beta-structure and its function is still
unknown.
III. Hydrolytic Activity
The
is an important feature of human salivary alpha amylase. The
loop is glycine-rich, malleable, and
holds the substrate in place during activity. It also
plays a role in the release of product. The loop is in
the "open" conformation when substrate is unbound, and
is in the "off" position when a substrate is attached. The
loop becomes more flexible and "opens" when it is time for
the products to be released. Then the cycle restarts.
When a carbohydrate binds to the active
site (located in Domain A), the flexible loop becomes more
structured and as a result moves into contact with the
substrate.
His201 first detects
the the substrate and causes the loop to take action. After
the loop binds to the substrate, several residues play
important roles.
Ala307 and
Gly306 protect the substrate with a hydrophobic
cover, while residues
Trp203 and
Trp 284 (located on the
secondary binding site) assist in hydrolytic activity.
Trp59 and Tyr62
are involved in stacking to bind glucose and guide it to the
catalytic residues.
Several
residues in Domain A assist in the binding and hydrolytic activity to
substrates.
Trp58 is critical for substrate binding, while
Trp59
and Tyr62 are
involved in stacking to bind glucose and guide it to the
catalytic residues. His299
and His101 are
necessary for catalysis.
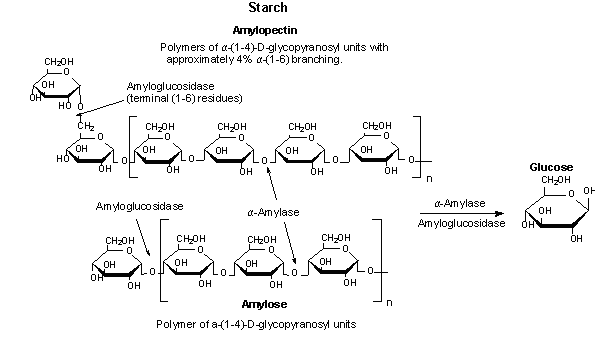
Figure 1. Alpha-amylase catalyzing the hydrolysis of starch to
glucose
IV. Bacterial Binding
Between
fluctuations in pH, temperature, nutrient supply, and saliva
flow, the oral cavity is a challenging location for bacteria
to persist. Human
salivary alpha-amylase binds to oral streptococci
in a similar fashion as it does to complex
carbohydrates. When breaking down bacteria,
His201
and
His305 are
necessary
to detect bacterial and starch substrates, while the flexible
loop attaches and
changes conformation, holding the substrate in place. Trp58
is vital for optimal hydrolytic activity.
It detects substrates and is thought to
assist in the orientation of the
substrate, as well as assist His305
to
its correct position.
V. References
1. Ramasubbu, Narayanan, Chandran
Ragunath, Prasunkumar J. Mishra, Leonard M. Thomas, Gyo¨
Ngyi Gye´ma´nt, and Lili Kandra. "Human Salivary A-amylase
Trp58 Situated at Subsite ) 2 Is Criticalfor Enzyme
Activity." Wiley Online Library. FEBS, 4 June
2004. Web. 02 Dec. 2015.
2. Hsiu, Julia, Edmond H. Fischer, and Eric A. Stein.
"Alpha-Amylases as Calcium-Metalloenzymes. II. Calcium and
the Catalytic Activity." Biochemistry including
Biophysical Chemistry & Molecular Biology. ACS
Publications, n.d. Web. 02 Dec. 2015.
3. Ramasubbu, Narayanan, Chandran Ragunath, Krishnan
Sundar, Prasunkumar J. Mishra, Gy¨ongyi Gyémánt, and Lili
Kandra. "Structure-function Relationships in Human
Salivary alpha-amylase: Role of Aromatic Residues." Academia.
Academia, 2005. Web. 02 Dec. 2015.
4.Lebenthal, E. (1987). Role of salivary amylase in
gastric and intestinal digestion of starch. Digestive
Diseases and Sciences, 32 (10), 1155-1157.
5. Butterworth, Peter J., Frederick J. Warren, and Peter
R. Ellis. "Human Alpha-amylase and Starch Digestion: An
Interesting Marriage." Wiley Online Library, 5 Jan. 2011.
Web. 02 Dec. 2015.
6. Nishide, T., Nakamura, Y., Emi, M., Yamamoto, T.,
Ogawa, M., Mori, T., Matsubara, K. (1986). Primary
structure of human salivary alpha amylase gene. Gene,
41(2-3), 299-304.
7. "3BLK." RCSB PDB. PDB-101, Worldwide PDB
Protein Data Bank, EMDataBank, Ndb, Structural Biology
Knowledgebase, n.d. Web. 02 Dec. 2015.
8. Ragunath, Chandran, Suba G.A. Manuel, Venkat
Venkataraman, Hameetha B.R. Sait, Chinnasamy Kasinathan,
and Narayanan Ramasubbu. "Probing the Role of Aromatic
Residues at the Secondary Saccharide Binding Sites of
Human Salivary alpha-amylase in Substrate Hydrolysis and
Bacterial Binding." Probing the Role of Aromatic
Residues at the Secondary Saccharide Binding Sites of
Human Salivary alpha-amylase in Substrate Hydrolysis and
Bacterial Binding 384.5 (2008): 1232-248. Journal
of Molecular Biology. U.S. National Library of
Medicine, 14 Oct. 2008. Web. 02 Dec. 2015.