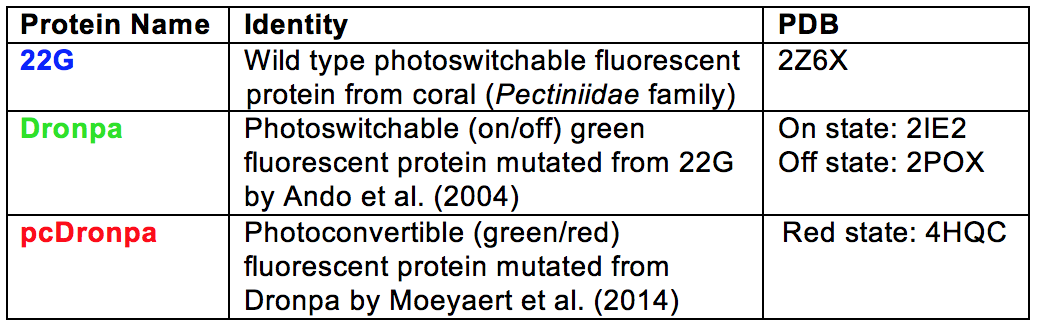
Dronpa: A Photoswitchable
Protein Derived from Coral (Pectiniidae)
Maria Sorkin '16 and Emily Bulik-Sullivan '16
Contents:
I. Introduction
Stony corals (class Anthozoa)
produce proteins that fluoresce
in the presence of ultraviolet (UV) radiation. The function of
these proteins is currently unknown, though it has been
speculated that they may help protect corals and their
zooxanthellae from superoxide radicals and the abundant UV in
shallow tropical waters [1-3]. In addition to their fluorescent
ability, these proteins tend to be small (approximately 238
amino acids), bright, and temperature- and pH-resistant. These
qualities make them desirable for use in a number of laboratory
applications, such as fluorescent tagging to observe protein
dynamics [4,5].
A drawback of using
fluorescent proteins is that they can often be visualized only
once before they are photobleached. To address this problem,
Ryoko Ando et al. created a mutant, named Dronpa*, of a
photoswitchable fluorescent protein from the Pectiniidae
family of coral. Dronpa's fluorescence results from a
three-residue structure called a chromophore, shown here in
the on state [4].
The
chromophore fluoresces green when exposed to 503 nm light and
is "turned off" by exposure to 488 nm light. Dronpa has two
unique and useful characteristics. First, the photoswitching
process is reversible; exposing off-state Dronpa to 405 nm
light converts
it back to its fluorescent (on-state) form [6,7]. The second
useful quality is that each protein can undergo photoswitching
over 100 times without photobleaching, a quality which has
many potential experimental applications [5].
Since the creation of
Dronpa in 2004, numerous groups have mutated the protein to
make it faster, more durable, and multicolored [8-10]. In this
tutorial, we first introduce the wild type protein, 22G, and
explore the mutations made to it in order to create Dronpa.
Next, we explain the mechanism by which photoswitching occurs,
examining differences in the on- and off-state Dronpa
structures. Lastly, we highlight a particularly useful Dronpa
mutant, named pcDronpa, that has green-to-red photoconversion
ability. In an attempt to clarify the different mutants from
the wild type protein included in this tutorial, each mutant
is represented by a different color scheme (see table below).
*After
the Japanese word dron
referring to ninjas disappearing and PA, for
photoactivation.
II. Original Mutation: 22G to Dronpa
The
wild type protein that Ando et al. originally purified from
coral, "22G," formed an oligomer with a molecular weight of
102 kDa. This was 3.5 times greater than the 29.2 kDa
molecular weight expected from the protein's primary structure
[4]. Ando et al.
therefore created a monomeric mutant ("22Gm3"
which they renamed Dronpa) with a molecular weight of 28.8
kDa. This monomer forms a tetrameric complex in vitro, as displayed in the "Show Wild Type Protein (22G)" button above.
To make Dronpa, Ando et al. introduced six
mutations to 22G: Ile102-Asn, Phe114-Tyr, Leu162-Ser,
Arg194-His, Asn205-Ser, and Gly218-Glu [4,10].
These
mutations break down the quaternary structure of the
tetrameric wild type protein to yield the monomeric mutant,
while also avoiding the chromophore to preserve the
protein's fluorescence.
Quaternary structure stabilization in 22G is demonstrated by
interactions between
Ile102+Ile102
and between Gly218
and residues including Pro141
[6]
.
It therefore follows that elimination of Ile102 and Gly218
contributes to tetramer dissociation.
III. General Structure of On-State Dronpa
In the on-state, Dronpa
crystals consist of four identical beta-barrel protomers, or
beta-cans, that each contain an interior chromophore [6].
The secondary structure of Dronpa is similar to that of other
fluorescent proteins. Each beta-barrel consists of 224 amino
acids, comprising numerous elements. Eleven beta-sheets
of differing lengths shield the chromophore from external
interactions. One of these, beta7, is divided into two
beta-sheets (beta7a and beta7b) by two amino acids. In
addition to the beta-sheets, each subunit of Dronpa has a
chromophore, two short alpha
helices between residues 54-59 and 77-82 that reach
into the beta-can to support the chromophore, and numerous linkers
that connect all of these elements.
The chromophore tripeptide Cys62-Tyr63-Gly64
(CYG) confers fluorescent function to the protein and sits inside a
hydrophobic pocket made up of Gln38, Met40, Thr58, Ile195, Leu209,
and Glu211.
CYG is held in its cis-
conformation in the beta-can by covalent peptide bonds with Phe61
and Asn65. Multiple
hydrogen bonds and van der Waals interactions further stabilize
the chromophore moiety.
Unlike most fluorescent proteins,
the Dronpa chromophore is attached from both ends to the beta-can
via co-axial alpha helices.
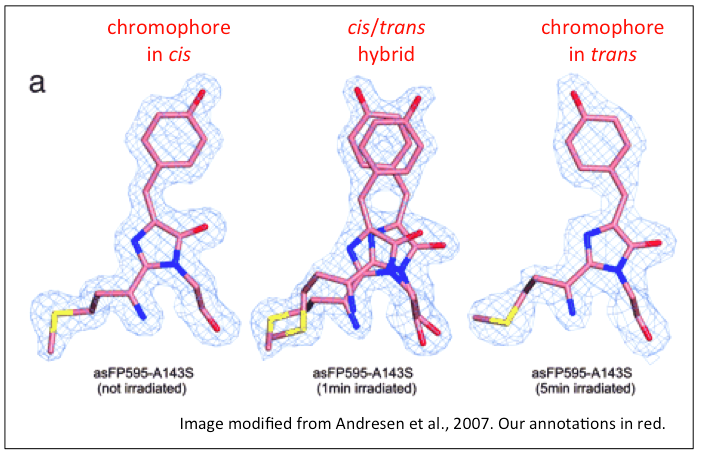
IV. Off-State
Photoswitchability back and forth from the fluorescent state
to the dark state is dictated by chromophore conformation.
When the chromophore tripeptide absorbs light of wavelength
488nm, it transitions from the cis- (fluorescent, or
"on state") conformation to the trans- ("off state")
conformation (see image below) [6,7].
Upon absorption of 405nm light, the chromophore switches back
to the cis-
conformation,
effectively restoring the fluorescence of the protein [9].
The off-state Dronpa protein is nearly identical to the
green-fluorescent state, with the exception of changes in
amino acid residues Arg-66,
Ser-142,
Val-157, and His-193,
which accommodate the transition from the cis-
conformation of the chromophore to the trans-
conformation. Slight rotation of these four residues results
in the rearrangement of the p-hydroxyphenyl ring
in the chromophore.
Movement of Ser-142 breaks a critical hydrogen bond to the p-hydroxyphenyl
ring, slightly destabilizing the chromophore.
Destabilization of the chromophore in the dark
state contributes to its inability to fluoresce.
V. Green-Red Photoconvertible Mutant
One mutant of important
experimental significance that has been created from
Dronpa is a green-red photoconvertible protein, named
pcDronpa. Photoconvertibility of a reporter protein
like Dronpa affords research groups the ability to use
multimodal imaging, using the two different
fluorescent colors to obtain high
resolution images that show precise localization
and activity of a target protein. Multimodal imaging
is being adapted to many existing technologies, such
at PET-CT scanners [11]. The rapid photoswitching
mutant can cut down experiment duration, making it an
appealing alternative to the original Dronpa protein.
PcDronpa was
created by making four mutations to the original Dronpa
mutant: Cys62-His,
Asn94-Ser,
Asn102-Ile, and Glu218-Gly.
The Cys62-His
mutation affects the chromophore itself, contributing to the
ability of pcDronpa to photoconvert. Note that the mutations
to amino acids 102 and 218 that enabled the original Dronpa
mutant to exist as a monomer have been reversed in pcDronpa,
permitting it to take on its tetrameric form
[8].
Similarly to the original Dronpa
mutant, this protein is switched to the dark state by
absorption of 488 nm light and fluoresces green by
absorption of 405 nm light. To convert to the red state,
high intensities of 405 nm light are required. Unlike the
green state, the red state is unable to photoswitch from the
on- to the off-state. Rather, conversion to the red state
cleaves the protein backbone, rendering it unable to switch
to an off state. The slightly non-planar characteristic of
the chromophore in the red state protein is the most notable
physical difference between the red and green protein
structures and is likely caused by a hydrophobic interaction
between the imidizole group (CH2NCH) of the chromophore
and Met40 [8].
VI. References
1. Banaszak AT, Lesser
MP. Effects of solar ultraviolet radiation on coral reef organisms.
Photochem Photobiol Sci. 2009;8:1276-1294. doi:10.1039/b902763g
2. Bou-Abdallah F, Chasteen ND, Lesser M. Quenching of Superoxide
Radicals by Green Fluorescent Protein. 2006;1760:1690-1695.
doi:10.1016/j.bbagen.2006.08.014
3. Eyal G, Wiedenmann J, Grinblat M, D'Angelo C, Kramarsky-Winter E,
Treibitz T, et al. Spectral Diversity and Regulation of Coral
Fluorescence in a Mesophotic Reef Habitat in the Red Sea. PLoS One.
2015;10:e0128697. doi:10.1371/journal.pone.0128697
4. Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic
shuttling observed by reversible protein highlighting. Science.
2004;306:1370-1373. doi:10.1126/science.1102506
5. Habuchi S, Ando R, Dedecker P, Verheijen W, Mizuno H, Miyawaki A,
et al. Reversible single-molecule photoswitching in the GFP-like
fluorescent protein Dronpa. Proc Natl Acad Sci U S A.
2005;102:9511-9516. doi:10.1073/pnas.0500489102
6. Wilmann PG, Turcic K, Battad JM, Wilce MCJ, Devenish RJ, Prescott
M, et al. The 1.7 Å Crystal Structure of Dronpa: A Photoswitchable
Green Fluorescent Protein. J Mol Biol. 2006;364:213-224.
doi:10.1016/j.jmb.2006.08.089
7. Eisenstein M. New fluorescent protein includes handy on-off switch.
Nat Methods. 2005;2:8-9. doi:10.1038/nmeth0105-8
8. Moeyaert B, Nguyen Bich N, De Zitter E, Rocha S, Clays K, Mizuno H,
et al. Green-to-red photoconvertible dronpa mutant for multimodal
super-resolution fluorescence microscopy. ACS Nano. 2014;8:1664-1673.
doi:10.1021/nn4060144
9. Stiel AC, Trowitzsch S, Weber G, Andresen M, Eggeling C, Hell SW,
et al. 1.8 Å bright-state structure of the reversibly switchable
fluorescent protein Dronpa guides the generation of fast switching
variants. Biochem J. 2007;402:35-42. doi:10.1042/BJ20061401
10. Kaucikas M, Fitzpatrick A, Bryan E, Struve A, Henning R, Kosheleva
I, et al. Room temperature crystal structure of the fast switching
M159T mutant of the fluorescent protein dronpa. Proteins Struct Funct
Bioinforma. 2015;83:397-402. doi:10.1002/prot.24742
11. Moseley, M, Geoffrey D.
Multimodality imaging. Stroke. 2004;35;2632-2634. doi:10.1161/01.STR.0000143214.22567.cb
12. Andresen, M, Stiel, AC, Trowitzsch, S, Weber, G, Eggeling, C,
Wahl, et al. Structural basis for photoswitching in Dronpa. Proc Natl
Acad Sci U S A. 2007;104:13005-13009. doi:10.2210/pdb2pox/pdb