Mtr4 RNA Helicase
Elizabeth Abrash '17 and Ulises Arbelo '16
Contents:
I. Introduction
Model View:
The Saccharomyces cerivisiae mRNA transport protein
(Mtr4) is a RNA helicase that interacts with the RNA exosome. The
exosome core is responsible for processing and degrading RNA.
In vivo function of the core requires several cofactors, on
of which is Mtr4. The RNA exosome core is conserved in
eukaryotes and Archaea. In both cases the central channel of the
exosome is known to be 8-10A wide, or wide enough to process
single-stranded RNA. As an RNA helicase, Mtr4 is, in part,
responsible for unwinding double stranded mRNA, so that it can be
degraded.
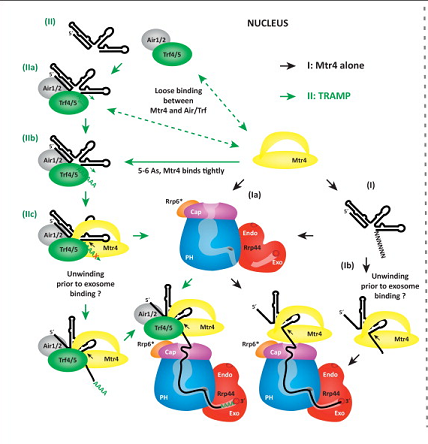
Mtr4 can act alone to unwind RNA, or it can interact with the
Tr4/5-Air1/2-Mtr4 polyadenylation (TRAMP) complexes. This complex
consists of a poly(A)polymerase (Tr4/5) and a Zn-knuckle RNA-binding
protein (Air1/2). In order to act on its own, Mtr4 requires a 3'
overhang of ~5-6 nt. If the RNA substrate does not have this
overhang, then it first binds the TRAMP complex until there is a
minimal binding site for Mtr4.
II. General Structure
Model View:
Mtr4 includes a DExH core formed by two RecA domains, the N
terminal beta hairpin, the Winged Helix domain, and the helical
bundle domain. The
, RecA-1 and RecA-2, bind RNA and ADP. The
, packs with the beta strands of RecA-1 and latches on to
RecA-2. RecA-2 is connected to the
, by 15 residues and the WH domain packs against the
. Mtr4 also features the
in between the wings of the winged helix.
III. The DexH Core
Model View:
The RecA domains of Mtr4 consist of central beta-sheets
surrounded by alpha-helices. This crystal structure shows eight beta
sheets in the RecA-1 domain and seven beta sheets in the RecA-2
domain.
The cleft between the two RecA domains contains eight conserved
motifs that are known to be involved in nucleotide and nucleic acid
binding, as well as ATP hydrolysis.
The RecA domains are stabilized by the N-terminal beta hairpin on
one side and by the helical bundle domain and C-terminal tail on the
other side. Conserved, hydrophobic residues in the C-terminus
interact with the cleft between RecA-1 and RecA-2. A fourteen
residue loop of the winged helix domain extends across RecA-1 in
order to connect RecA-2 to the winged helix.
IV. The Stalk/Kow Insertion Domain
Model View:
The Stalk/Kow insertion domain is a 270 residue insert
between the third and fourth helices of the Winged Helix domain.
It begins with alpha helices alpha-1 and alpha-2 of the Stalk
perpendicular to each other, continues into the KOW
(Kyrpides-Ouzounis-Woese) region and finishes with helices alpha-3
and alpha-4, which wrap around the first two alpha helices.
The KOW region seems to be a flexible unit, while
the flexibility of the Stalk region seems to be restricted by
intramolecular contacts at the hinge regions. The arch-like
feature of this protein is largely due to a 120 degree turn made
in the hinge region, which is likely due to the presence of
several conserved residues.
facilites the bend, which is stabilized by hydrophobic packing
of several conserved
residues: Val637, Val643, Tyr646,
Leu840, Leu846, and Tyr853.
Two invariant residues, Glu640 and Lys856 are near each other
due to the sharp bend in the hinge region. The best explanation
for the absolute conservation of these two residues is an
interaction between the two. It is believed that these residues
are oriented in a way that would allow a
to form.
V. ADP and RNA Binding
Model View:
ADP binds at the cleft between the two RecA domains which are
conserved across DExH proteins. The ADP is recognized by Gln154 which is characteristic of the
Q-motif
conserved across DEAD box helicases and forms hydrogen bonds
with the adenine amino group (Figure credit Tanner et al.,
2003). The ADP-Mtr4 binding is further stabilized by the
stacking of the adenine ring with Arg547
and Phe148.
Like ADP, RNA also binds to the RecA-1/2 cleft. The helicase
activity of Mtr4 occurs at the conserved beta-hairpin formed at residues
521-532. In particular, the RNA interacts with
Trp524 and Gly526 which
unwinds the double stranded RNA. The separation is further aided
by electrostatic interactions between
Lys 523 and Arg530
and the single stranded RNA.
While not the main RNA binding site, it is possible that
conformational changes in the
affect RNA binding affinity. Mtr4 mutants lacking the Stalk/KOW
domain have shown reduced RNA binding as well as lower ATPase
activity.
VI. Trf4-Air2 Recruitment
Model View:
The Stalk/KOW domain was originally seen as potential TRAMP
binding site since both are specific to Mtr4. It also
hypothesized that the polyadenylated RNA might be fed to the
Mtr4, and thus the TRAMP might be located near the RNA binding
site. Nonetheless, mutants of Mtr4 missing the Stalk/KOW
domain had similar TRAMP binding as wild-types indicating that
the DexH core, again, is the relevant binding site.
Further studies identified several key
sections of the RecA domains involved in TRAMP
binding (Figure credit Falk et al., 2014). The Trf4 Asp122
forms
with Arg349 and Asn352 . Trf4 binding is further
strengthened by a
edge-to-face
interaction between Trf4 Phe123
and Mtr4 Tyr339.
Air2 has several
interactions along the RecA2 domain. Air2 Tyr45
participates in
with Mtr4 Ile464, Leu479, and
Leu483. Nearby, Air2 Arg44
forms
with Glu1021 and a cation-pi
with Tyr1020 .
Air2 also has a helix secondary structure which
forms hydrogen bonds with the Mtr4 helical bundle
domain at Lys1015
and hydrophobic interactions with
Met1016
.
VII. References
Falk S, Weir JR, Hentschel J, Reichelt P, Bonneau
F, Conti E. 2014. The Molecular Architecture of
the TRAMP Complex Reveals the Organization and
Interplay of Its Two Catalytic Activities. Molecular
Cell Vol 55, 6 : 856-867
Jackson, RN, Klauer AA, Hintze BJ, Robinson H,
Hoof AV, Johnson SJ. 2010. The crystal structure
of Mtr4 reveals a novel arch domain required for
rRNA processing. The EMBO Journal Vol 29,
13
Schneider, C, Tollervey D. 2013. Threading the
barrel of the RNA exosome Trends in
Biochemical Sciences Vol 38, 10: 485-493
Tanner, KN, Cordin O, Banroques J, Doere M, Linder
P. 2003. The Q Motif: A Newly Identified Motif in
DEAD Box Helicases May Regulate ATP Binding and
Hydrolysis. Molecular Cell Vol 11, 1 :
127-138
Weir, JR, Bonneau F, Hentschel J, Conti E. 2010.
Structural analysis reveals the characteristic
features of Mtr4, a DExH helicase involved in
nuclear RNA processing and surveillance. PNAS
107:27, 12139-12144
Back to Top