E. coli MGMT Suicide
Protein
Ben Canniff '19 and David Anderson '19
Contents:
I. Introduction
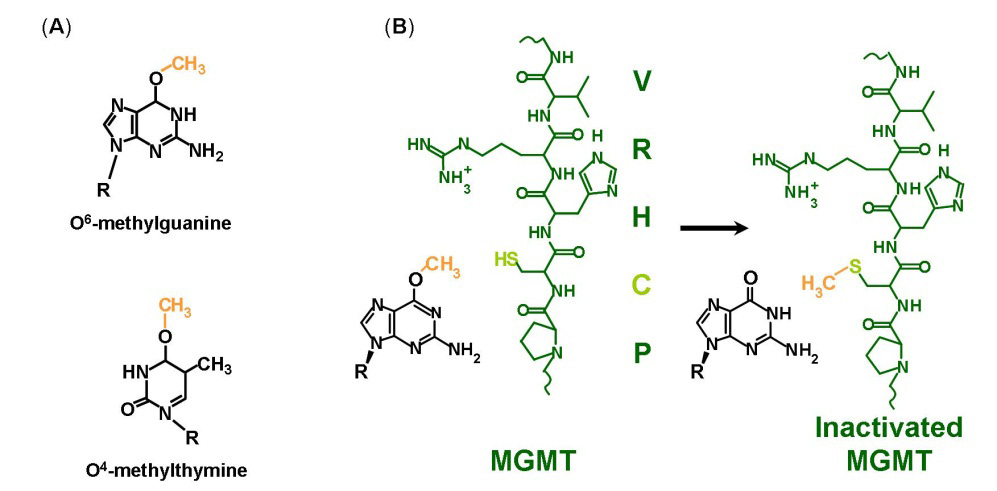
The Escherichia coli Ada gene O6
–methylgunine-DNA methyltransferace (MGMT) is a DNA binding protein
that is involved in repairing mutations that occur during DNA
replication. MGMT participates in methylation, which means it is a
protein that adds a methyl group. MGMT adds a methyl group onto a
specific cysteine residue, Cys146
, by breaking an ether bond connecting the methyl to the O6 guanine
within the DNA chain. By simultaneously binding the methyl group
onto itself and breaking the methyl's old bond, MGMT is rendered
permanently inactive which is why it has earned the title of "The
Suicide Protein".
The repair protein is located in the nucleus to allow it to
have the best access to the newly replicated DNA. The alkyl groups
targeted by MGMT are direct results from alkylating mutagens and
carcinogens that cause mistakes in the replicated DNA. These
mistakes must be fixed before the DNA replicates creating a mutant
daughter strand.
(L., Stephanie, and Timothy R 2013)
II. General Structure
The suicidal DNA repair protein Ada O-6-methylguanine-DNA
methyltransferase (MGMT) is 354 amino acids in length and 39 kDa in
molecular mass. Ada is unique in the fact that it is present as a
monomer in both its native and methylation state. The protein is
predominantly composed of alpha helixes
along with a group of beta sheets
(Moore et al. 1996).
MGMT’s structure allows for stoichiometric alkyltransferase
action at two different substrates. The action at different
substrates is able to occur because MGMT contains two different
functional domains, the 10 kDa N-terminal
and the 19 kDa C-terminal
(Moore et al. 1996).
The N-terminal domain acts as a phosphotriester-DNA,
methyltansferase to accept alkyl groups from the diastereoisomers of
alkylphosphotriester residues. The C-terminal domain, termed Ada-C,
accepts alkyl groups at
from O6 alkylguanine and O4 alkylthymines. The alpha helixes suggest a
"helix-turn-helix" motif
within the C-terminal Domain where MGMT contacts DNA. The Escherichia
coli MGMT protein contains
the Pro-Cys-His
-Arg
active site sequence contained in an alpha helix that is shared between all known organisms
possessing an MGMT protein (Moore et al. 1996).
III. DNA Binding
MGMT’s native structure does not allow for the active site
to interact with the target O6 methyl group. In order for the protein
to correct a mutation on the DNA strand, it swivels one of the
C-terminal helices to expose a possible DNA binding site. The
swiveling of the C-terminal breaks the His147
and Glu
173 hydrogen bond
allowing the His147 to rotate so it, along with the rest of the active
site, faces the incoming DNA molecule. (Bhattacharyya et al. 1998) Cys
146 is the active site amino acid that performs a
methytransferase action by while the helix-turn-helix
motif contacts the major groove of the DNA. (Moore et al. 1996)
Cys 146
carries out a necleophilic attack on the target methyl group ,
which is allowed by a conformational change of the protein when a
methylguanine
is detected. This conformational change is required in order for
MGMT to activate. Alkylation slightly distorts the phosphate-sugar
backbone of dsDNA while MGMT shows a strong preference for dsDNA
over ssDNA.
The C-terminal active site helix attaches to the major groove of the
DNA, covering around 8 base pairs of
DNA.
MGMT does not repair methylguanine in Z-DNA because its left hand
helical orientation does not allow for MGMT to contact the DNA
correctly (Reinhard et al. 2001). Z-DNA
Overview of DNA Binding
IV. Methy Transfer Reaction
The MGMT protein removes mutagenic methyl groups from
guanines and thymines through the methyltransferase activity.
MGMT binds the methyls to the sulfur
molecule in Cys-146 located within the C-terminal.
In order to
transfer the methyl from the O6-methylguanine to the Cys-146,
the protein must undergo acid catalyzed SN2 chemistry, which
allows MGMT to bind the methyl while simultaneously cleaving the
ether bond. While SN2 chemistry takes place a thiolate, an
organosulfur compound, acts as a nucleophile. During the
reaction a naturally associated positive charge from MGMT
transfers to the O6 guanine and the carbon center undergoes a
nucleophilic attack by the thiolate, thereby breaking the O 6
guanine’s ether bond to the DNA and binding it to the thiolate,
forming a thioether, and the deactivated Cys-146
residue. Once the thioether bond is made the protein undergoes
ubiquitination and is degraded. (Moore et al. 1996)
VI. References
Moore, M.h., J.m. Gulbis, E.j. Dodson, B.
Demple, and P.c.e. Moody. "Ada O6-Methylguanine-Dna
Methyltransferase From Escherichia Coli." (1996): n. pag. Web.
Bhattacharyya, Debasish, Tapas K. Hazra,
W. David Behnke, Parkson L.-G. Chong, Alexander Kurosky, J.
Ching Lee, and Sankar Mitra. "Reversible Folding of Ada
Protein ( O 6 -Methylguanine?DNA Methyltransferase) of
Escherichia Coli †." Biochemistry 37.6 (1998): 1722-730. Web.
Reinhard, Jost, William E. Hull,
Claus-Wilhelm Von Der Lieth, Uta Eichhorn, Hans-Christian
Kliem, Bernd Kaina, and Manfred Wiessler.
"Monosaccharide-Linked Inhibitors OfO6-Methylguanine-DNA
Methyltransferase (MGMT): Synthesis, Molecular Modeling,
and Structure?Activity Relationships." Journal of Medicinal
Chemistry 44.24 (2001): 4050-061. Web.
L., Stephanie, and Timothy R. "Direct
Repair in Mammalian Cells." New Research Directions in DNA
Repair (2013): n. pag. Web.
Silva, Nathan, and David Marcey. "An
Introduction to Jmol* Scripting** Nathan Silva and David
Marcey © 2016." Intro to Jmol Scripting. N.p., 2016. Web. 07
Dec. 2016.
Back to Top