Human Cytochrome P450 3A4
Wyatt Cole '19 and Tim Lewis '18
Contents:
I. Introduction
Cytochromes P450 are a family of enzymes, which play critical
role in the body by metabolizing endogenous compounds, and many
xenobiotics or substances foreign to the body. Cytochrome P450 34A
(CYP3A4) is one isoform of Cytochromes P450. CYP3A4 is a heme
containing enzyme which metabolizes many marketed drugs. Cytochromes
P450's many isoforms are responsible for the metabolism of more than
90% of marketed drugs, and CYP3A4 metabolizes more marketed drugs than
all the other P450 isoforms combined.Despite CYP3A4’s critical role in
drug metabolism our understanding of how it recognizes these varying
chemical structures has been limited, without an accurate
three-dimensional structure. Researchers have previously been forced
to use homology models sharing less than 25% sequence identity with
CYP3A4. Williams et al. seek to correct this problem by reporting the
crystal structure of both unlinganded CYP3A4 and CYP3A4 bound to
either the inhibitor metyrapone of the substrate progesterone.
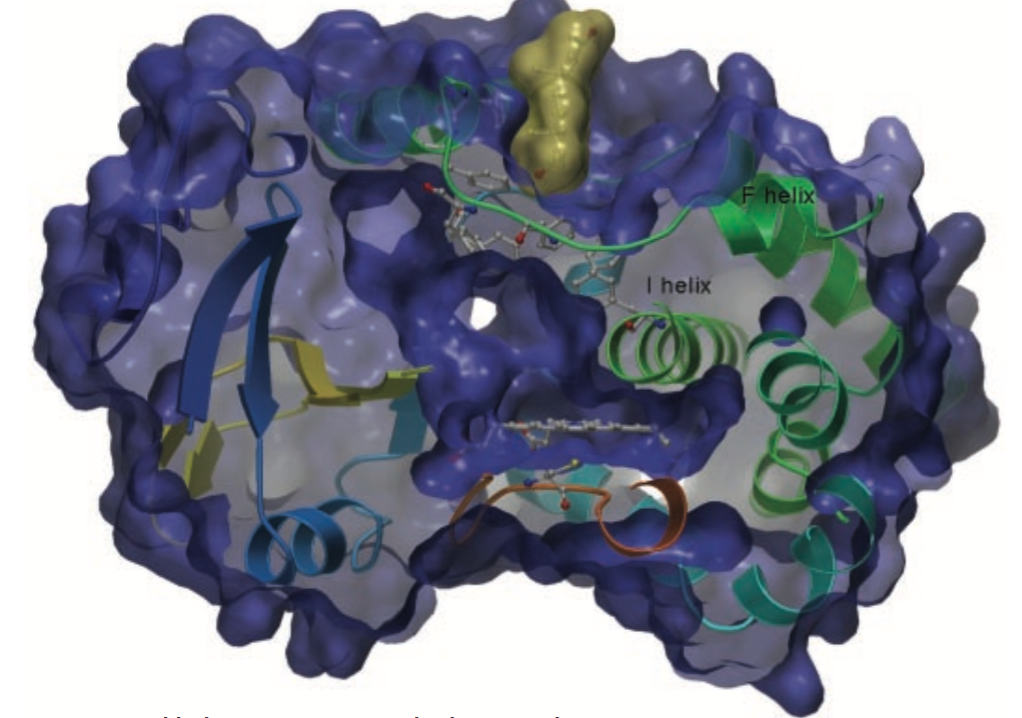
II. General Structure
CYP3A4 conforms to many of the patterns seen in P450’s many
isoforms such as a small N-terminal domain and a larger helical
C-terminal domain. The C-terminal domain contains the enzymes active
site with the
. This heme iron is ligated by a conserved cysteine Cys-442 and
interacts with Arg-105, Trp-126, Arg-130, Arg-375, and Arg-440
.There are two channels in this protein one formed by the
and the other formed by the
. The B’ and C helices separate these channels; this region can change
its conformation depending on the presence or absence of a ligand.
CYP34A also has several unique features such as a hydrophobic region
located around the loop following the
.
CYP3A4 also has a strikingly small helix without any secondary
structure, in the region following
.CYP3A4 also has a
, consisting of Phe-108, Phe-213, Phe-215, Phe-219, Phe-220 , Phe-241
and Phe-304, located above the active site and forming a hydrophobic
core through pi stacking. Changes in CYP3A4 have been shown to occur
in the presence of a substrate effector molecule bound in the active
site. The active site of CYP3A4 lies in the helical C-terminal
domain and is composed of three subpockets. This active site is
surprisingly small particularly when the large diversity of CYP3A4
substrates is taken into account. This small active site is
predicted to undergo substantial conformational changes including
the movement of the Phe cluster and the
to accommodate larger substrates. It is also predicted that movement
of the
region could also open up the channel and increase the volume of the
active site.
III. Metyrapone and Progesterone Binding
Metyrapone’s binding results in no fundamental protein
conformational changes. The crystal structure of CYP34A shows
metyrapone bound by an alkyl-pyridine nitrogen to the heme iron.
Metyrapone is a small substrate and its binding in the active site
leaves room for other molecules to potentially bind. Progesterone
also induced very little conformational change binding at a
peripheral site close to the Phe cluster and away from the heme
iron. Progesterone nestles against the Phe cluster on the side
chains of
and interacts with the protein through a hydrogen bond between its
acetyl oxygen and the amide nitrogen. Although progesterone could
potentially bind the active site of CYP3A4 there is no evidence that
this actually occurs. Progesterone’s binding in the peripheral
binding pocket is supported by the residues,
, implicated in progesterone’s homotrophic and heterotrophic
cooperativity existing in the vicinity of this pocket.
Peripheral binding of Progesterone to
CYP3A4
IV. Ketoconazole
Binding
The large antifungal drug ketoconazole induces dramatic
conformational changes in CYP3A4. The Crystal structure determined
by Ekroos et. al. shows
bond in the active site. The keto group of the first ketoconazole
molecule is located in the polar pocket, lined by the side chains
, Ketoconazole binding is further stabilized in CYP3A4 by chlorobenzyl
ring pi-stacking with the side chain of
. The other ketoconazole molecule is stacked in an antiparallel
orientation above the first. This ketoconazole’s keto group
hydrogen bonds with the side chain of
. This simultaneous binding of two ketoconazole ligands could be an
artifact of high concentrations of ketoconazole used in
crystallization. There were notable conformational changes
observed in the F and G helices and intervening loops upon
ketoconazole binding. In ketoconazole-CYP3A4 complex the
is also broken up with some of its hydrophobic chains exposed to the
surrounding medium. When ketoconazole is bound Arg-212, which is
usually found in CYP3A4’s active site is found on CYP3A4’s
surface,
which is usually found on the protein surface is found in the active
site. There are also distortions seen in the loop connecting the
C-terminal and beta-sheets and in the I helix cleft when
ketoconazole is bound.
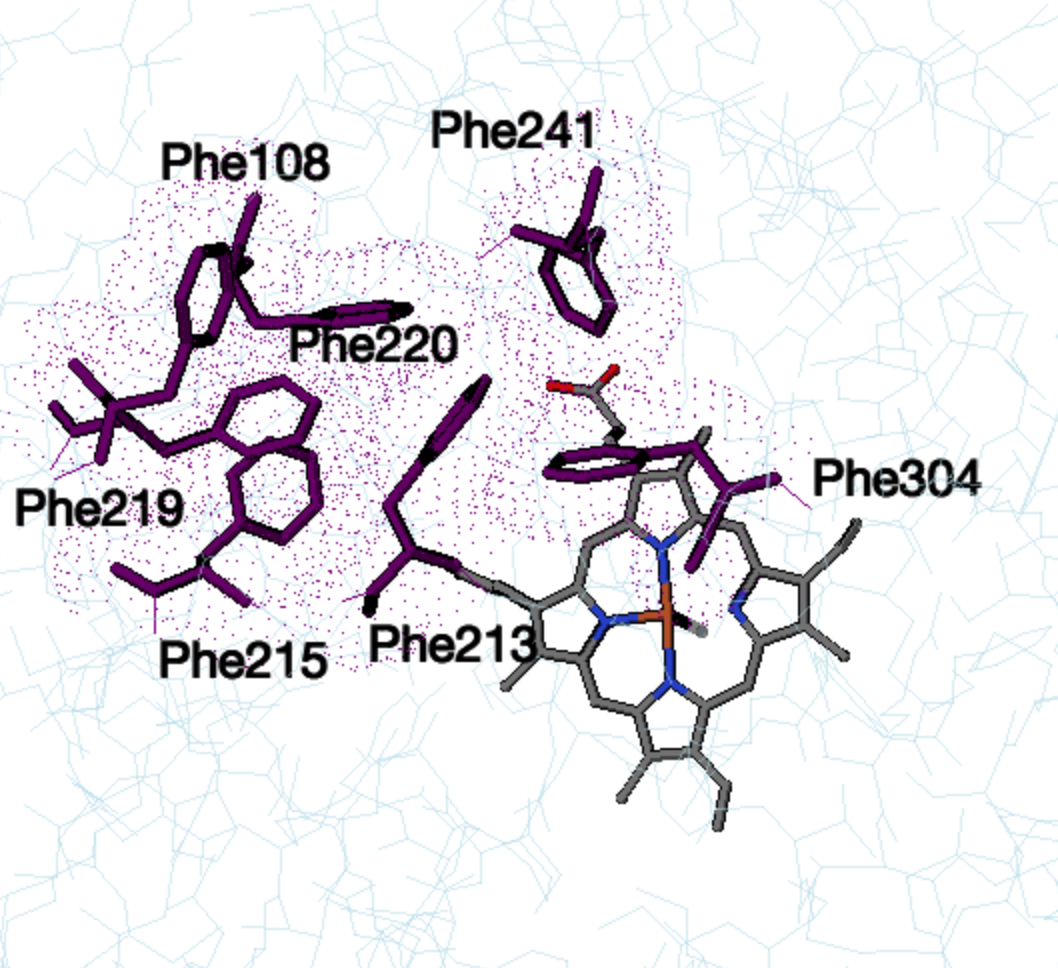
Phe cluster in unbound CYP3A4
VI. References
Ekroos M, Sjogren T. Structural basis for
ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci USA.
2006; 103(37):13682-13687. DOI:10.1073/pnas.0603236103
Williams AP, Cosme J, Vinkovic´ DM, Ward A,
Angrove H, Day P, Vonrhein, Tickle I, Jhoti H. Crystal
Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and
Progesterone. 2004; 5684:683-686. DOI: 10.1126/science.1099736
Poulos T, Finzel BC, Howard AJ.
High-resolution Crystal Structure of Cytochrome P450cam. J. Mol.
Biol. 1986;195;687-700. PMID: 365642828:4568-4574.
Back to Top