Exoribonuclease Rat1 and
Activating Partner Rai1
Ty J. Boyd'20, Peter E. Reinhart'20
Contents:
I. Introduction
Introduction: 5’ to
3’ exoribonucleases are a large family of conserved enzymes in
eukaryotes. These enzymes have important functions in RNA metabolism
and interference. Schizosaccharomyces pombe Rat1,
or XRN 2 in humans, also plays a major role in the termination of
RNA Polymerase II. Rat1
is stimulated by its activating partner, Rai1.
In vitro, Rat1 is
unstable and loses nuclease function upon pre-incubation unless in
complex with Rai1.
The Rat1-Rai1
complex forms a more stable secondary structure, allowing for the
efficient degradation of RNAs. Independent of its interaction with Rat1, Rai1
has pyrophosphohydrolase activity toward the 5’ triphosphorylated
end of RNA. This type of activity is crucial for degradation of
mRNA.
II. Rat1
and Rai1 Structure
Rat1 Active
Site
S. pombe Rat1
has two conserved segments. These two sequences make up a large domain
of the protein
. Of these conserved
sequences, the first segment
makes up the majority of the enzyme’s active site. This region shares
has several weak nuclease homologs in the PDB (i.e RNase H). Like
these other nucleases, the active site is made up of a similar cluster
of acidic residues. Suggesting that these proteins share the same
catalytic activity. The second
segment surrounds the active site in a "pocket-like fashion"
not found in other homologs, making few direct contributions active
site.
Xiang et al, Figure 1c
The unique pocket-like structure is responsible for Rat1’s
inability to perform endonuclease activity. Thus, providing a
Molecular explanation for the exclusivity of its exonuclease activity.
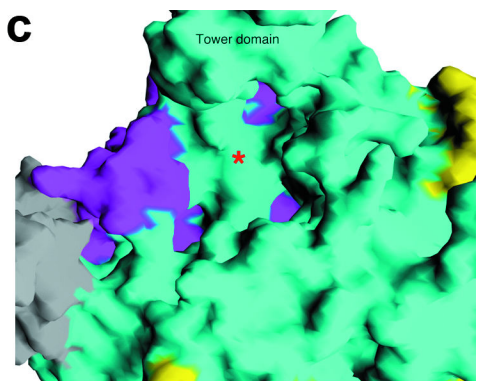
Tower Domain
Rat1
contains a notably large helix (αD) that extends 30Å out from the
protein. This structure is known as the
of Rat1.
While the C-terminal of the tower extends far from the protein, the
N-terminus contains several conserved residues to the active site.
Interestingly, Xrn1 homologs have deletions in the C-terminus of the
enzyme, suggesting that tower domain may serve functions that are
specific to Xrn2 such as Pol II termination.
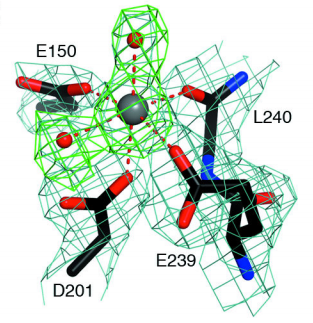
Rai1 Structure
Structures of Rai1
and its homolog DOM3Z contain two “highly twisted” β-sheets and
several nearby helices. This motif, along with the unique sequences
of these proteins would suggest a new protein fold. More
importantly, this structure may suggest catalytic function. The
residues of in
addition to the main chain carbonyl of Leu240 and two water
molecules form the coordination sphere of a cation.
Xiang et al, Figure 3c
The pocket here serves as an active site for Rai1
and its homolog. Interestingly, the conformation of the active site is
almost identical in DOM3Z, despite the protein not interacting with Rat1. This suggests the
active site functions independently of the binding of Rat1.
Rat1-Rai1
Interface
Rai1 is bound 30Å
away on the face opposite to Rat1’s
active site. The main interactions involve the C-terminal segment
of Rat1 and the
β8-αE segment of Rai1
(as well as strand β4). Mutations in the residues of in Rai1 greatly reduced
interactions between the Rat1
and Rai1 . Mutation
of Rat1 residues on
the interface had similar consequences.
III.Rai1
Pyrophosphohydrolase Activity
Data from exoribonuclease assays show that the catalytic
activity of
Rai1
like that of a 5’ RNA pyrophosphohydrolase. This activity turns a
5’-triphosphate group into a 5’-monophosphate group, thus allowing
the RNA to become a substrate for
Rat1.
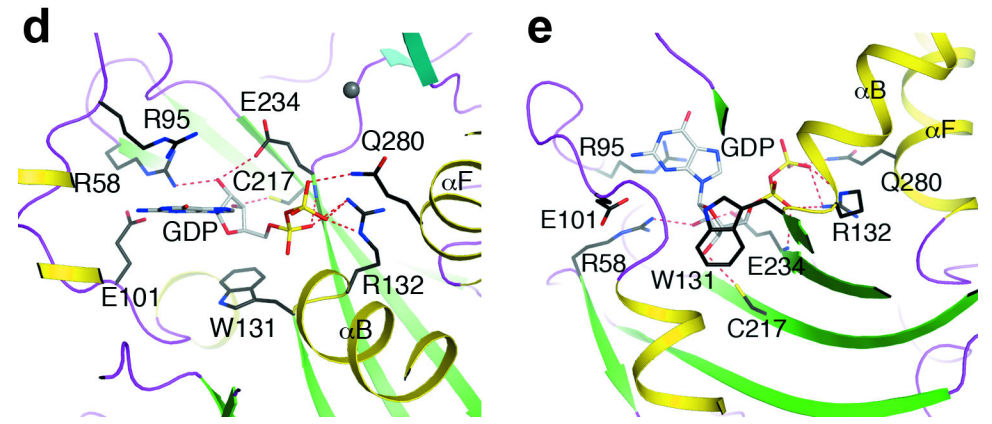
Interactions between GDP and the [DOM3Z active site],
Rai1
analog, have helped to define the substrate binding mode of the
active site of
Rai1.
The phosphate of GDP makes strong interactions with the dipole
of helix αB near the N-terminus. Direct interactions are also made
with the side chain of Arg94(DOM3Z Arg132). The β phosphate makes
interactions with the same Arginine as well as Gln263(DOM3Z
Gln280) in the αF helix. Interestingly, the hydroxyls as well as
the guanine base are not recognized by any conserved residues,
though the ribose does interact with Trp93 (DOM3αZ Trp131) via Van
der Waals. Finally, conserved Gln199 (DOM3Z Glu234) is located
near the phosphates of GDP and may serve a catalytic role in the
of
Rai1.
Xiang et al, Figure 3d,e
IV.Termination of RNAPII by Rat1
One of the most important roles of
Rat1
is its function in the termination of RNA Polymerase II (RNAPII)
during transcription. Loss of
Rat1(or
Rai1) has been shown
to result in defective and inefficient transcription termination.
The
torpedo
model of transcription termination suggests that
Rat1 (XRN2) enters a
polyA cleavage site and degrades the nascent RNA until eventually
displacing RNAPII from DNA. More specifically, it has been
suggested that the length of the nascent RNA degraded by
Rat1
is correlated positively with the overall efficiency of RNAPII
termination. This suggests that, not only does the exoribonuclease
activity of
Rat1
allow it to gain access to the polymerase, but it may also play a
role in developing a “driving force” in order to displace the
polymerase. Interestingly,
Rat1’s
inability to terminate the RNAP in
E. coli suggests
interactions between the polymerase and
Rat1
must maintain a certain level of specificity. These interactions
are not yet well understood due to a lack of structural studies of
the
Rat1-RNAPII
complex.
V. References
1) Xiang, S., Cooper-Morgan, A.,
Jiao, X., Kiledjian, M., Manley, J. L., & Tong, L.
(2009). Structure and function of the 5’?3’
exoribonuclease Rat1 and its activating partner Rai1.
Nature, 458(7239), 784–788.
http://doi.org/10.1038/nature07731;
2)Nagarajan, V. K., Jones, C. I., Newbury, S. F., & Green, P. J. (2013). XRN 5’?3’ exoribonucleases: Structure, mechanisms and functions. Biochimica et Biophysica Acta, 1829(0), 590–603. http://doi.org/10.1016/j.bbagrm.2013.03.005
3)Park, J., Kang, M., & Kim, M. (2015). Unraveling the mechanistic features of RNA polymerase II termination by the 5?-3? exoribonuclease Rat1. Nucleic Acids Research, 43(5), 2625–2637. http://doi.org/10.1093/nar/gkv133
4)Lemay, J.-F., & Bachand, F. (2015). Fail-safe transcription termination: Because one is never enough. RNA Biology, 12(9), 927–932. http://doi.org/10.1080/15476286.2015.1073433Lemay, J.-F., & Bachand, F. (2015). Fail-safe transcription termination: Because one is never enough. RNA Biology, 12(9), 927–932. http://doi.org/10.1080/15476286.2015.1073433
Back to Top