STAT6 Transcription Factor
Hannah L. Hertz '19 and Meredith K. Glover '20
Contents:
Model View:
I. Introduction
The JAK-STAT pathway transmits extracellular signals (cytokines and
growth factors) from the cell membrane to the nucleus. Malfunctions in
this pathway are known to result in immune system disorders and cancers.
The JAK-STAT pathway is composed of three main components: a cell
membrane receptor, a Janus kinase (JAK), and two Signal Transducer and
Activator of Transcription (STAT) proteins. Although the mammalian STAT
family consists of seven functionally related proteins with conserved
domain organization— STAT1, 2, 3, 4, 5a, 5b, and 6— only STAT6 and STAT3
play major roles in the JAK-STAT pathway. [1]
Here, we focus on STAT6, a transcription factor
primarily stimulated by two cytokines: interleukin (IL) 4 and IL-13.
Under normal conditions, when stimulated by IL-4 and IL-13, STAT6 is
phosphorylated, forming a homodimer that translocates to the nucleus (Fig.
1). In the nucleus, STAT6 acts as a transcription factor,
activating genes responsible for the differentiation of T helper 2
cells. [2-3]
Mutations in the transcription factor STAT6 may result in disease.
For example, STAT6 is associated with asthma development. Furthermore,
STAT6 is constitutively active in many types of cancers. Thus,
targeting the functioning of STAT6 is an attractive therapy for many
diseases.[4]
In order to target STAT6, a better understanding of STAT6
transcriptional regulation is needed.
Li et al. (2016) examine the molecular details on how
STAT6 recognizes and binds specific segments of DNA. With this
molecular biology tutorial, we hope to illustrate the structural basis
of STAT6 DNA recognition and binding as outlined by Li et al. (2016).
Only the core fragment of STAT6 is shown in this tutorial (residues
123-658), which binds to DNA with affinities comparable to full length
STATs. [4]
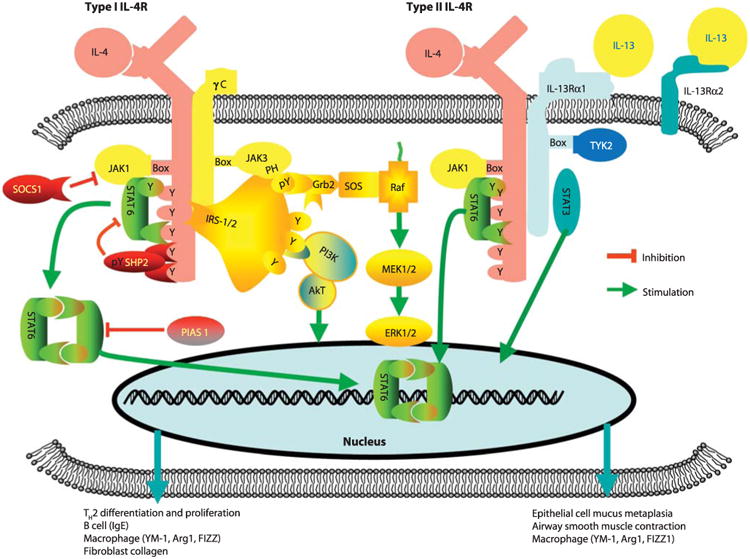
Figure 1. The JAK-STAT pathway. STAT6 is indicated
in green. (Wills-Karp and Finkelman, 2008)
II. General Structure
STAT6 is a homodimer (composed of two identical
),
capable of binding DNA when phosphorylated. STAT6 is made up of
: the N-terminal coiled coil
domain (residues 130-274), the DNA
binding domain (DBD) (274-443), the Linker
domain (443-536), the SH2
domain (536-626), and the C-terminal phosphotyrosine
tail segment (626-651).
How does STAT6 form a homodimer? Phosphorylation of the residue
occurs at the phosphotyrosine tail segment of both monomers. The amino
acids in the phosphotyrosine tail
segments interact with the amino acids in the SH2
domain of the adjacent monomer via hydrogen
bonds
; the hydrogen bonds hold two identical STAT6 monomers together,
forming the STAT6 homodimer. Dimerization of phosphorylated STAT6
forms an
.[4]
III. STAT6 vs. Other STAT Structures
The structure of STAT6 differs slightly from other STAT homodimers.
The STAT6
(aa 609-620) of the SH2 domains are shorter than the STAT1 and STAT3
C-terminal loops. This causes an
to form above the antiparallel beta-sheet. Furthermore, the four
of the STAT6 coiled coil domain are shorter than the corresponding
regions in STAT1, STAT3, and STAT 5a.
Another key difference between STAT6 and other STATs is the angle of
dimerization. In STAT6 (either in complex or not in complex with DNA),
the angle of intersection of the homodimer is larger than the angle of
intersection formed in the STAT1 and STAT3 homodimers. These
differences allow the dimeric assembly of STAT6 to be more flexible
and thus more accessible for DNA binding in comparison to other STATs.
The structural basis of STAT6 DNA binding also differs; only one
residue of STAT6 (in comparison to three residues in STAT1) forms a
bond with a DNA base. This interaction is explored further in the next
section. [4]
IV. DNA Binding
Only phosphorylated STAT6 forms a homodimer capable of DNA binding.
Dimerization of phosphorylated STAT6 forms the antiparallel
beta-sheet. Two
located on the antiparallel beta-sheet
serve as hinge axis points around which the two molecules of STAT6
rotate when the dimer binds to DNA. Truncations of STAT6 before
lose the ability to bind DNA due to the loss of intermolecular
interactions of the antiparallel beta-sheet and phosphorylated tail
segments, and thus, reduced dimerization.
How does the STAT6 transcription factor locate its target genes?
Conserved DNA motifs are found in the region of STAT6 target genes.
STATs recognize a DNA motif (5'-TTCN3/4GAA-3’
) with conserved palindromic bases (TTC/GAA)
seperated by a spacer consisting of 3 (N3) or 4 (N4) nucleotides of
any of the four nucleotides found in DNA. STAT6 is the only STAT
protein that prefers binding to DNA motifs with a N4 spacer.
One STAT6 homodimer binds a palindromic DNA motif. Each monomer
binds half of the palindromic sequence. Residues at
in the DBD participate in the recognition and binding of the
palindromic DNA motif. The residues in the DBD bind DNA via hydrogen
bonds with the DNA backbone
.
is the only residue that forms hydrogen bonds with a DNA base--the O6
of guanine at position 14 in the N4 DNA motif. H415 is oriented
outward from the STAT6 homodimer, and thus available for DNA binding.
Li et al. (2016) confirm that H415 has an important role in
recognizing DNA motifs with a N4 spacer. Mutations of H415 decreased
STAT6's binding affinnity to 5'-TTCN4GAA-3’
and increased STAT6's binding affinity for 5'-TTCN3GAA-3’.
Upon DNA binding, the monomers of STAT6 undergo a significant
rotation. This rotation shifts the position of H415 along with placing
the DBD at an optimal binding position. The STAT6 dimer becomes more
compact once bound to the N4 DNA motif. [4]
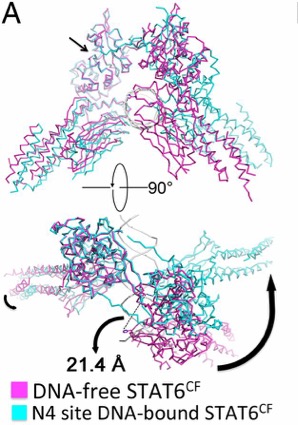
Figure 2. Conformational change of STAT6 upon DNA
binding. Black arrows show movement. (Li et al., 2016)
V. Disease-Associated Mutations
Mutations in STAT6, especially in the DNA binding loops, are
associated with many diseases. Abnormalities in the STAT6 DNA binding
loop can lead to asthma, follicular lymphoma
,
or cancer. Li et al. (2016) hypothesize that key mutations in the
STAT6 DNA binding loop are associated with disease because these
mutations decrease the electronegativity of the DNA binding interface
thus increasing the DNA binding affinity of STAT6. With a better
understanding of the mechanism of STAT6 DNA binding, we may develop
personalized medicine against STAT6 associated disease. [4]
VI. References
[1] Rawlings JS, Rosler KM, Harrison DA. (2004).
The JAK/STAT signaling pathway. Journal of Cell Science.
117: 1281-1283.
[2] “STAT6 Gene (Protein Coding).” Gene Cards
Suite, Gene Cards Human Gene Database, 3 Dec. 2017, www.genecards.org/cgi-bin/carddisp.pl?gene=STAT6.
[3] Wills-Karp M and Finkelman FD. (2008).
Untangling the Complex Web of IL-4 and IL-13 Mediated Signaling
Pathways. Science Signaling. 1 (51): pe55.
[4] Li J, Rodriguez JP, Niu F, Pu M, Wang J, Hung
LW, Shao Q, Zhu Y, Ding W, Liu Y, Da Y, Yao Z, Yang J, Zhao Y, Wei
GH, Cheng G, Liu ZJ, Ouyang S. (2016) Structural basis for DNA
recognition by STAT6. PNAS. 113 (46): 13015-13020.
Back to Top