CLOCK:BMAL1
Brad Clegg '20 and Hannah Sklar '20
Contents:
I. Introduction
Model View:
Many important physiological and developmental processes have
basic helix-loop-helix PER-ARNT-SIM (bHLH-PAS) proteins as critical
regulators of gene expression networks, but most of these structures
are very poorly characterized. In order to form a functional DNA
binding domain, these bHLH proteins must dimerize, with the bHLH-PAS
proteins forming a specific dimer that is conferred by their PAS
homology domains (Kewley et al., 2004).
The mammalian circadian clock runs by an autoregulatory
transcriptional feedback mechanism. The bHLH-PAS proteins, CLOCK
and BMAL1, play a key
role in this mechanism as transcriptional activators (Huang et
al., 2012). CLOCK
and BMAL1 drive
cadenced gene expression by rhythmically activating the expression
of their respressors, PER and CRY, enhancing the transcription of Period
and Cryptochrome genes during the daytime (Menet et al.,
2014). At night, the proteins coded for by these genes, PER and CRY,
build up, dimerize, and translocate into the nucleus to associate
with the CLOCK:BMAL1
heterodimer, repressing further transcription. The PER:CRY repressor
is eventually broken down by ligase complexes, allowing the CLOCK:BMAL1
complex to stimulate transcription again, starting the new circadian
cycle. This reiterative process in the mammalian circadian clock
repeats roughly every 24 hours.
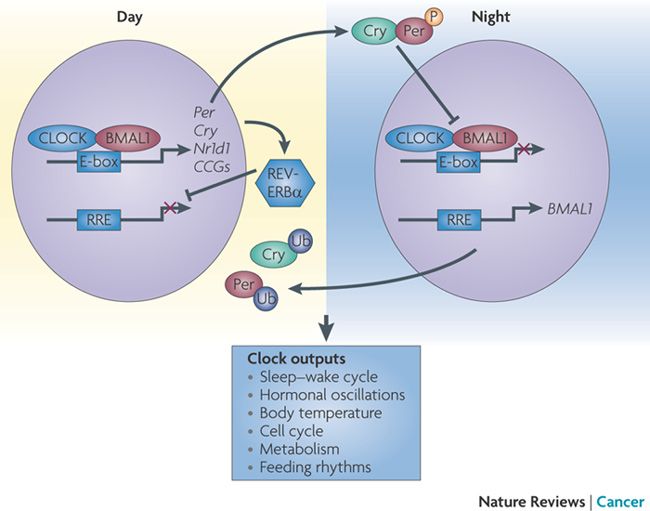
Figure 1: Repressors PER and CRY interact with
CLOCK:BMAL1 heterodimer at night and not during the day, creating the
circadian rhythm in mammals (Sahar and Sassone-Corsi, 2009).
II. General Structure
In order to obtain a protein complex stable enough for
crystallographic analysis, Huang et al. used protein
constructs containing two tandem mouse BMAL1
(PAS-A and PAS-B)
and CLOCK
(PAS-A and PAS-B)
and a
domain for each subunit (BMAL1
bHLH and CLOCK bHLH).
Looking at the three-dimensional structure of the CLOCK:BMAL1
complex, there is a tightly intertwined heterodimer with the three
domains in CLOCK
interacting with their partner domains in BMAL1.
Although the three domains have very comparable amino acid sequences
with their respective partner domain in the other subunit, there are
drastic conformational differences between the different domains in
their spatial relationship. This makes it so that this is an
unusually asymmetric heterodimer.
This asymmetry is shown in multiple structural properties of
the heterodimer, with one being that the BMAL1
bHLH alpha-2 helix is arranged
with the A'alpha helix of the BMAL1
PAS-A domain. This is very different in the CLOCK subunit,
which has a
between the end of the alpha-2 helix of bHLH
and the start of the A'alpha helix of PAS-A,
and is connected by a flexible linker.
This results in direct contact between PAS-A
and the alpha-2 heix of bHLH in
CLOCK, while there is no direct contact between the same domains in
BMAL1.
The asymmetry is also shown by the electrostatic potential of
the two subunits. BMAL1
has an overall positive electrostatic potential while that of the CLOCK
subunit is overall negative. This is consistent with the exposed
faces of the subunits, since the exposed regions of BMAL1
display a positive or neutral electrostatic potential while CLOCK
has faces with a negative electrostatic potential. This source of
asymmetry adds an important property governing the potential
intereactions of the dimer and explains why the PER1, PER2, CRY1,
and CRY2 proteins can bind differentially with the CLOCK
and BMAL1 subunits.
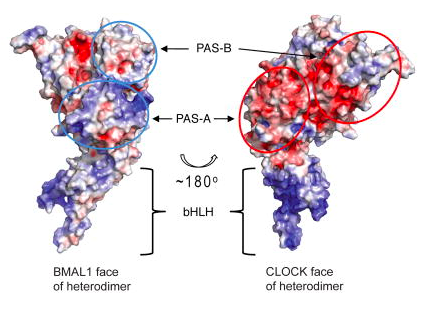
Figure 2: Electrostatic potentials of the exposed
faces of the CLOCK:BMAL1 heterodimer. Positive potentials are
indicated by blue ovals and negative potentials are indicated by red
ovals (Huang et al., 2012).
III. DNA Binding
In order for the heterodimer to act as a transcriptional
activator, it must be able to bind DNA. In order to confirm this,
the researchers used oligonucleotides containing th E-box sequence
5' CACGTG 3' from the mPer1 and mPer2 promoters and
verified that binding to the protein complex occurred (Huang et
al., 2014). Perhaps the most important part of the heterodimer
in terms of DNA binding is the
formed with the C-terminal halves of the alpha-1 helices
collectively with the alpha-2 helices of both CLOCK
and BMAL1 bHLH domains. Like in
other bHLH proteins, dimerization of the complex helps to stabilize
the hydrophobic core of this bundle. In order to recognize the E-box
DNA, it is also essential for the bHLH domains to be in the correct
conformation. To ensure precise interaction with the major groove
sites of the E-box DNA duplex, the alpha-1 helices must be exactly
aligned. The importance of this interaction is shown through a
mutation of the hydrophobic core
CLOCK L57 and L74, BMAL1
L95 and L115 to E, which completely abolished the transactivation
activity of the full length CLOCK:BMAL1 mutants (Huang et al.,
2014). It has been shown that neither subunit alone binds DNA well
without dimerization, so mutations that decreased dimerization also
decreased DNA binding.
IV. Dimerization
When forming the CLOCK:BMAL1
heterodimer complex, each domain interacts with the corresponding
domain of the other subunit (For example: CLOCK bHLH with BMAL1
bHLH). One of the most important components of the interaction
between the bHLH subunits in formation of the heterodimer is the
hydrophobic core in the four-helical bundle, with mutation leading
to a great reduction in the ability to form a stable heterodimeric
complex.
Interaction between the PAS-A domains of the subunits was
shown to be mainly facilitated by hydrophobic interactions. This
is primarily from
of the A'alpha helix of CLOCK
with the beta-sheet face of BMAL1.
Specific residues involved in this interaction were shown to be
in CLOCK and
in BMAL1, with double mutation to
E and D respectively making association between the subunits
undetectable (Huang et al., 2012).
On the PAS-B domains, the
most involved in dimerization are the CLOCK
helical face and the beta-sheet
face of BMAL1. It was shown that altered PAS-B domain
interactions resulted from mutations to CLOCK
W284, V315 and BMAL1 V435,
decreasing dimerization of the proteins. There was a significant
decrease in dimerization when mutating BMAL1
, significantly altering the hydrophobic interactions. One of the main
residue contacts involved in the interaction of the PAS-B domain
is between
. The double mutation of these residues to A led to a decrease in
dimerization as well as transactivation activity (Huang et al.,
2012). This confirms the unusual PAS-B domain association
involving the CLOCK helical face
and the BMAL1 beta-sheet face
as seen in the crystal structure.
V. Role in Circadian Cycling
CLOCK and BMAL1
are able to regulate the circadian clock in mammals through
physical interaction with regulators PER and CRY. The specifics
of this interaction are not known on a structural basis, but the
binding of these repressors could affect DNA binding, change
transactivation potential, or even reshape interactions with
other activators or repressors. It has been shown that
overexpression of CLOCK
leads to a shorter circadian cycle while overexpression of BMAL1
either lengthens or does not affect the cycle.
Research suggests that CRY interacts with the PAS-B
domain of CLOCK and with a C-terminal region of BMAL1.
Explicit amino acid residues involved in the interaction of the
CLOCK PAS-B domain in
repression by CRY are
. These residues are located on the solvent exposed HI loop of the
beta-sheet face and are readily available for interaction with
the repressor. CRY binding to CLOCK
is also consistent with the fact that CRY is a highly positively
charged protein and the exposed surfaces of CLOCK
have negative charges.
Interaction of BMAL1
with the PER repressor is not as well defined. Structural
similarities between the tandem PAS domains of the proteins show
possible structural or functional conservation. Further, the
residue located at the HI
loop of the BMAL1 PAS-B domain (which corresponds to
CLOCK W362, involved in association with CRY) is conserved in
Drosophila and mouse PER proteins and is involved in the
formation of homodimers with a second PER protein. This suggests
the importance of a conserved W residue at this position
mediating protein-protein interactions.
VI. References
Huang, N., Chelliah, Y., Shan, Y., Taylor,
C., Yoo, S., Partch, C., Green, C., Zhang, H., and Takahashi,
J. (2012). Crystal Structure of the Heterodimeric CLOCK:BMAL1
Transcriptional Activator Complex. Science, 337(6091),
pp. 189-194
Kewley, R., Whitelaw, M., and
Chapman-Smith A. (2004). The Mammalian Basic
helix-loop-helix/PAS Family of Transcriptional
Regulators. The International Journal of
Biochemistry and Cell Biology, 36(2), pp. 189-204
Menet, J., Pescatore, S., and
Rosbash, M. (2014). CLOCK:BMAL1 is a Pioneer-like
Transcription Factor. Genes and Development,
28:8-13.
Sahar, S. and Sassone-Corsi, P.
(2009). Metabolism and Cancer: the Circadian Clock
Connection. Nature Reviews Cancer, 9:886-896.
Back to Top