Crystal Structure of the
Glucocorticoid Receptor Ligand Binding Domain Reveals a Novel Mode
of Receptor Dimerization and Coactivator Recognition
Edwin Ramirez '20 and Nicole Steady '21
Contents:
I. Introduction
Nuclear receptors are a group of proteins found in cells that
regulate expression of genes that affect the development and
homeostasis of the cell through sensing different types of steroids,
hormones and other types of molecules. The
is a type of nuclear receptor found in humans that
can regulate: bone turnover, cell differentiation, lung maturation,
inflammation, etc.[1]. As found in Vareen et al. (1998), the
glucocorticoid receptor can regulate gene expression via:
glucocorticoid/GR complexbinding directly to specific DNA sequences
and regulating or activating certain genes, affecting the mRNA
stability, and through interactions with transcriptional factors.
This receptor is very often targeted by common ligands such as
dexamethasone and other glucocorticoid analogs which can treat can
treat medical conditions, such as the ones listed before but the
protein is most commonly targeted to treat inflammatory diseases
such as asthma.
II. Mechanism
There is a myriad of GR isoforms, but the two most common
isoforms of the GR are the alpha and the beta GR, which can be found
in the lining of the bronchioli of the lung. The GR is composed of
three main domains, the N-terminal activation function 1-domain
(AF-1), the central DNA binding domain (DBD), and the C-terminal
ligand binding domain (LBD). The DBD bind to the DNA at the
Glucocorticoid Responsive Elements, which is a promoter region on
the DNA. The dimer can bind positively with a palindromic consensus
sequence pf GGTACAnnnTGTTCT to cause transcriptional activation, or
negatively where the binding sequence can vary to cause repression
of a gene. Glucocorticoids can also regulate gene expression through
an enhanced mechanism that allows for ribonucleases to degrade
specific mRNA that contains AU sequences in the untranslated 3’
region. The final way in which glucocorticoids regulate gene
expression is interactions between the GR/glucocorticoid complex and
transcriptional factors such as AP-1 and NF-kB which are used to
mediate the induction of inflammatory genes. These protein-protein
interactions result in downregulation of receptors for these
transcriptional factors.
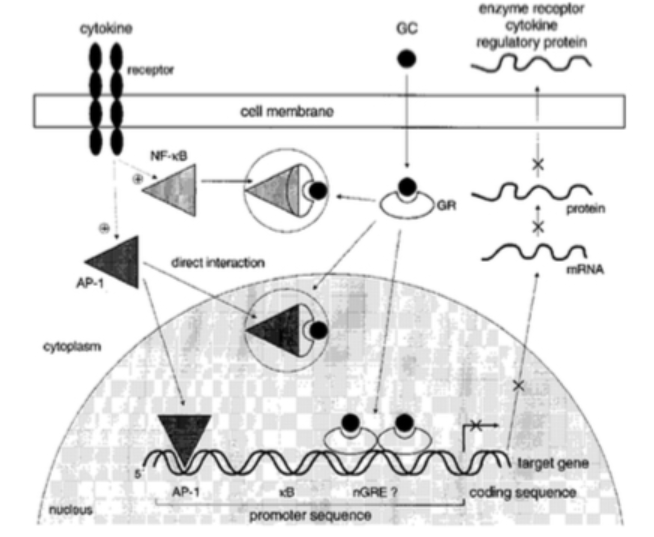
Velden et al. (1998)
III. C-Terminal Ligand Binding Domain
The composition of the GR LBDmonomers forms a
that is stabilized by a series of
, in an antiparallel conformation, formed by residues
547-551 of helices 1 and 3 from each LBD. The structure of the LBD
involves 11 alpha helices and 4 beta strands that make up a
three-layer helical domain. Helices 1 and 3 for the top layer, while
helices 7 and 10 construct the bottom layer, and, lastly, helices
4,5,8, and 9 form the middle layer but are found closer to the top
layer than the bottom layer.[1] The space that is left open in the
bottom half of the helical sandwich contains the hydrophobic pocket
to which a ligand,
, can bind to. The
contains reciprocal interactions between residues
P625 and I628 of beta strands 3 and 4 of the LBD. The LBD also
contains a C-terminal
which activates the LBD by creating a conformational
change by packing together the helices 3,4 and 10. The AF-2 helix is
stabilized by an
that forms a
with the beta strand of helices 8 and 9.
IV. Dexamethasone and TIF2 Recognition
Most coactivators such as
, bind to a nuclear receptor through three LXXLL
motifs, but most NR have had only two out of the three motifs
crystallized in complex. Bledsoe et al discovered the third motif of
the GR and TIF2 complex to be
, which also forms a two-turn alpha helix that
orients the hydrophobic leucine side chains into a groove. The N-
and C-terminal ends of the coactivator helix are stabilized by the
E755 residue of the AF-2 helix and the K57 residue of helix 3 of the
GR LBD, which serves as a
. Bledsoe et al all support the presence of a
between residues D590 and R585 of the GR and +2 and
the +6 sites of the LXXLL motif. When in the hydrophobic pocket, the
ligand has both hydrophobic and hydrophilic interactions with
helices 3,4,5,6,7,10, AF-2, and residues from beta strands 1 and 2.
Dexamethasone has its A ring facing towards Beta strands 1 and 2,
and its D D ring facing towards the AF-2 helix
. With this orientation comes an extensive number of
and
with almost every atom in dexamethasone which
ultimately helps to stabilize the ligand in this binding center.
Lastly, the ligand makes
with residue L753 of the AF-2 helix, and the
residues I747 and F749 of the loop that proceeds the AF-2 helix in
order to keep the AF-2 in the active binding site conformation.
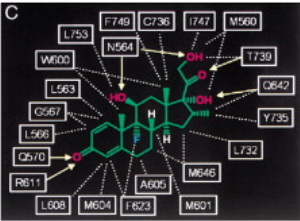
Bledsoe et al. (2002)
V. N-terminal Activation Function-1 domain and DNA Binding
Domain
The N-terminal can be as small as 24 amino acid residues or
as long as 600 amino acid residues and it is understood that this
component of the GR is significant for transcriptional activation.
AF-1 domain activates transcription by binding to sites on DNA that
are similar to those used for the DBD, GREs. The DBD has two zinc
ions that each contain different interactions with the DBD residues.
Both contain 4 cytosine interactions with the zinc ions, which
creates “zinc fingers”, but one is followed by the H1 helix, the DNA
reading helix, which contains an arginine residue that is used to
bind to specific bases of the major groove of the DNA, whereas the
other “zinc finger” is followed by the H2 helix which makes contact
to the minor groove of the DNA and contains peptide loop, “D loop”,
that is used to stabilize the dimer of the DBD. Both zinc fingers
are connected by “lever arm” loop.
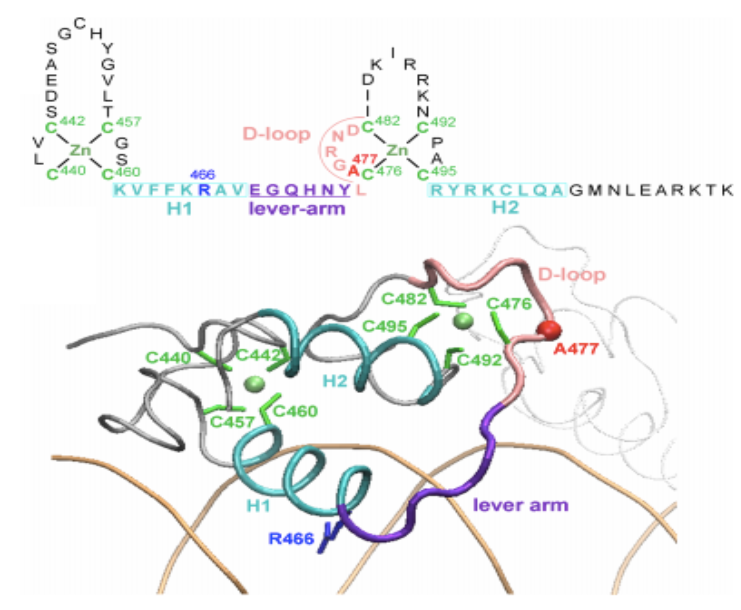
Burgess et al. (2017)
VI. References
Álvarez, L. D., Presman, D. M., & Pecci, A. (2017). Molecular dynamics simulations of the glucocorticoid receptor DNA-binding domain suggest a role of the lever-arm mobility in transcriptional output. PLoS ONE, 12(12), 1–18
Bledsoe, Randy K., Montana,Valerie G., Stanley,Thomas B.,
Delves,Chris J, Apolito, Christopher J., McKee, David D., Consler,
Thomas G., Parks, Derek J. Stewart, Eugene L., Willson, Timothy
M.,Lambert, Millard H., Moore, John T. 2002. Crystal Structure of
the Glucocorticoid Receptor Ligand Binding Domain Reveals a Novel
Mode of Receptor Dimerization and Coactivator Recognition.
Cell Press, Vol. 110: 93–105.
Burgess, A., Shah, K., Hough, O., & Hynynen, K. (2016). HHS Public Access, 15(5), 477–491. https://doi.org/10.1586/14737175.2015.1028369.Focused
Velden, V. H. J. van der. 1998.
Glucocorticoids: mechanisms of action and anti-inflammatory
potential in asthma. Mediators of Inflammation, Vol.7:
229–237.
Back to Top