Ku Recruitment for the
Repair of Double Stranded DNA Breaks
Tobias McCabe '21, Amna Tahir '21, and Jessica Meza '21
Contents:
I. Introduction
Molecule:
View Type:
When there is a double stranded break in the DNA (DSB), it is repaired
through the non-homologous end joining (NHEJ) pathway. This pathway requires many complexes such as
Ku, XRCC4 and DNA ligase IV to mediate the repair process. Although
the DSB repair through the NHEJ pathway does not use a template to
rejoin the separated ends of DNA, NHEJ is a highly accurate mechanism
of repair. Ku is responsible for coordinating the accurate alignment
and stabilizing of disjointed DNA strands. Proteins including
XLF and
APLF interact with Ku through a series of residues that make up Ku
Binding Motifs (KBM) in order to commence repair. When
X-KBM and
A-KBM
are bound to DNA, they can stabilize and rotate
Ku80 interactions with
the DNA so that repair may begin.
II. General Structure
The Ku
is made of two subunits
70K and
80K.
is located nearer to the N-terminus and
is near the C-terminus. There is a Ku heterodimer located on each side
of a DSB;
Ku70 is about 7 angstroms closer to the
broken portion of
DNA than
Ku80
because
Ku80 is largely responsible for
the binding of outside factors of the NHEJ pathway.
APLF and
XLF (and
other factors) each have their own KBM but compete for the hydrophobic
pocket of
Ku80.
III. DNA Binding
Ku forms a
ring like structure formed by
both dimers called the
.
That non-sequence specific
ring binds to
.
The
DNA binding domain has six Rossmann
fold B sheets and is 70 angstroms long. Although
Ku
binds to
DNA through interactions with
the sugar phosphate backbone,
also interacts with the minor groove.
DNA
is held securely in place due to a combination of positive
electrostatic charges along the inside of the
ring.
About two turns of
DNA, or 20 base pairs,
can fit inside of the
DNA binding domain
ring. Because the
ring
binding domain allows about 70% of the
DNA
to be exposed, repair factors are available to access the
DNA.
The
groove is located near to the N-terminus while the C-terminus is
important in the binding of the NHEJ factors.
IV. APLF interactions with Ku
interacts with a
of the Ku80 subunit. The KBM of APLF is highly conserved; in humans
the sequence is 5' - AERKRILPTWMLA - 3'
and varies slightly in other mammals. APLF binds at the edge of the
which is 50 angstroms away from Ku80-DNA binding site.
When interacting with vWA, the hydrophobic APLF adopts a hairpin
conformation and sits in the pocket of Ku created by helices a4 and a5
and the beta strands B and C. Residues 5'-
Leu68, Tyr74, and Ile112 - 3' are responsible for the tight
between the Ku pocket and A-KBM . The
basic residues in APLF's N-terminus (Glu181, Arg182, Lys183) and an
acidic residue (Arg184) form
through hydrogen bonds with Ku's Asp106, Asp109, Gln73, Ser145, Lys144, and Ser143.
V. XLF Interactions with Ku
When the Ku-h
DNA complex is bound to XLF peptides via
, there is a large outward rotation of the
Ku80
and the remainder of the Ku heterodimer. This formation of the large
groove is known as the "open" state of Ku. This conformational change
doesn't affect the way Ku interacts with double-stranded
DNA.
X-KBM binds near
the
Ku80 vWA face of this groove in the pocket that is
indicated by the position of four
(BA, BD, BE, and BE') and three
(a2, a7', a7").
X-KBM binds closer to the
DNA than the
APLF
but it isn't in direct contact. In the
, the residues PHE41, LEU234, VAL37, LEU12,PHE135, PHE164, and PHE225 are buried and mediate intermolecular contact. They occupy a
hydrophobic pocket which stabilizes the open state of
Ku80. The vWA
opening also has a secondary structure that separates the vWA from the
rest of the Ku heterodimer; it is found within the
(R232-E241). In the absence of
DNA, or with short
DNA that is
protected by the
Ku ring,
XLF and
X-KBM will interact similarly with
Ku because their binding affinities to
Ku are very similar. There is
also a 50 base pair interaction between
XLF and Ku.
Figure 1. from (1) Nemoz et al. The structural components of Ku and other factors.
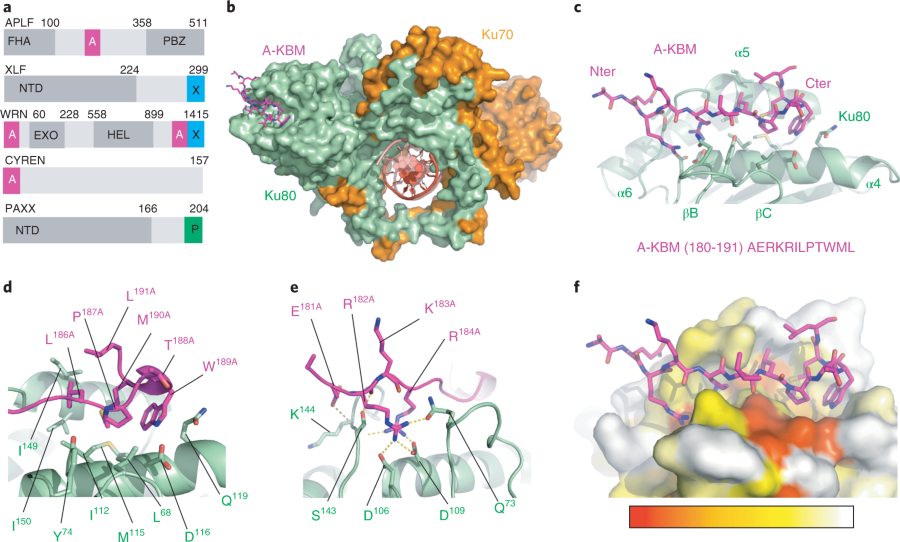
VI. Important Residues
is seen as a potential "spring" to help with the Ku80 opening
based on its position in the closed state of Ku80. It's pKa is 9.1
(typically in solvent, Glu is 4.5) which provides energy from the
solvation of the Glu E133 for Ku80 to open once the Ku80 residues,
V236, F237, and I240, are displaced away from the E13380 carboxyl
moiety. A patch of the N terminus, followed by a hydrophobic
patch, share similar sequences between A-KBM and X-KBM. A-KBM
affinity relies on
while X-KBM affinity relies on
X-KBM with an L297W substitution had an interaction 40-fold
tighter than a WT X-KBM whereras the L297E mutation had no
interaction with Ku. However, when saturated with A-KBM, X-KBM
L297W had no interaction with Ku, which suggests that the mutation
redirected X-KBM to the A-KBM binding site on Ku80. The X-KBM
fragment gets redirected to the APLF-binding site by the L297W
mutation. APLF-specific interaction with Ku80 is determined by the
W189 residue. WT X-KBM bound to the Ku-DNA complex was bound
stronger than with both the L297W and L297E mutations. This shows
that the L297W mutation cannot redirect the XLF protein to the
A-KBM binding site, even if X-KBM is redirected by the mutation,
leading to impaired recruitment of XLF.
VII. References
1. Nemoz, C.; Ropars, V.; Frit, P.; Gontier, A.;
Drevet, P.; Yu, J.; Guerois, R.; Pitois, A.; Comte, A.; Delteil,
C.; Barboule, N.; Legrand, P.; Baconnais, S.; Yin, Y.; Tadi, S.;
Barbet-Massin, E.; Berger, I.; Le Cam, E.; Modesti, M.;
Rothenberg, E.; Calsou, P.; Charbonnier, J. B. XLF and APLF bind
Ku80 at two remote sites to ensure DNA repair by non-homologous
end joining. Nature Structural & Molecular Biology 2018,
25, 971-980.
2. Walker, J. R.; Corpina, R. A.; Goldberg, J.
Structure of the Ku heterodimer bound to DNA and its
implications for double-strand break repair. Nature 2001,
412, 607.
Back to Top