EF-G in Complex with GDP and
the Ribosome in Thermus thermophilus
Zac LaRocca-Stravalle '21 and Kylie Writer '21
Contents:
I. Introduction
Model View:
Elongation Factor G (EF-G) is a prokaryotic translation factor involved
in the catalysis of translocation. As part of the
guanosinetriphosphatase (GTPase) superfamily, EF-G functions via
guanosine triphosphate (GTP) binding. In this complex, EF-G�GTP binds to
the ribosome and catalyzes the translocation of peptidyl-tRNA from the
aminoacyl (A) site to the peptidyl (P) site in the small subunit (30S)
of the ribosome. As this occurs, a single mRNA codon shifts 5� to 3�
relative to the ribosome. After translocation is complete, GTP is
hydrolyzed to guanosine diphosphate (GDP) and a conformational change
occurs in the EF-G complex. In this new conformation, the EF-G�GDP
complex dissociates from the ribosome and subsequently forms a complex
with GTP in a catalytic cycle (Al-Karadaghi et al., 1996).
The EF-G�GDP complex has important structural interactions in the
translocation process. The conformational change induced by GTP
hydrolysis is extensive in EF-G�GDP, and its interaction with the
ribosome also contribute to such changes. In this tutorial, we will
identify the basic components of EF-G as well as the major interaction
sites and structural changes with regard to GDP and the ribosome.
II. General Structure
The three dimensional structure of EF-G is presented to the left. In
this structure, EF-G is in the GDP complex conformation. EF-G is made
up of 691 amino acid residues with a molecular weight of ~80 kDa and a
length of ~110 �; it is the largest GTPase known (Jurnak et al.,
1994). The
of EF-G is made up of 19 alpha helices
(177 residues; 25%) and 42 beta sheets
(217 residues; 31%). EF-G can be broken into 5
: G domain (N-terminal), domain
II, domain III, domain
IV (C-terminal), and domain V .
The
: can be further divided into the G core
subdomain and the G insert subdomain
(G�); both GTP and GDP bind
at the G core (Czworkowskil, et al.,1994; Al-Karadaghi, et al., 1996).
All domains make contact with the ribosome. Both domain
II and domain III contact with
the 30S subunit of the ribosome, with further contact in the 50S
subunit by domain III. Domain
V primarily contacts the 50S subunit. Domain
IV, inserted into the 30S subunit, interacts with the P-site
tRNA and mRNA.
III. GDP Binding
binds to the G core subdomain at
the nucleotide-binding site. The two phosphates of GDP
interact with the main chain atoms in the
(residues 19-26), while the ribose sugar and guanine base interact with
the side chain residues in the G core
domain (Al-Karadaghi et al., 1996). The specific residues that
interact with GDP, via hydrogen bonding, are as follows:
:
Beta Phosphate:
Asp 22 (N) interacts with O23.
Gly24 (N) and Ala 23 (N) interact
with O21.
Thr26 (N) and Lys 25 (N) interact
with O22.
Alpha Phosphate:
Thr 27 (OG1 and N) interact with O13.
:
Lys 138 (N) interacts with O4'.
:
Asn 137 (N) interacts with N7.
Ser 262 (O) and Ala 263 (N)
interact with O6.
Asp 140 (OD1 and OD2) interact with N1 and N2
respectively.
IV. GDP Induced Conformational Changes
Studies comparing EF-G�GDP with EF-G in the nucleotide free state have
noted the local changes around the nucleotide-binding site. Upon
incorporation of GDP, a conformational
flip occurs in the P-loop, leading to the displacement of
(residues 116�123) and the
(residues 85�100) as well as in parts of domains III,
VI, and V.
The specific amino acid changes are described as follows: when GDP
enters the site, it forces the displacement of
in the P-loop. The
of IIe21 that would normally occupy the position of the beta
phosphate of GDP gets moved 5.4 �, 3.1 � from N delta 1 of
His20, in GDP occupancy. The
between the O delta 2 of Asp22 and N delta 1 of His20 is then replaced
by
to N zeta of Lys138. Additionally, Ile21 and Asp22 get rotated 180�.
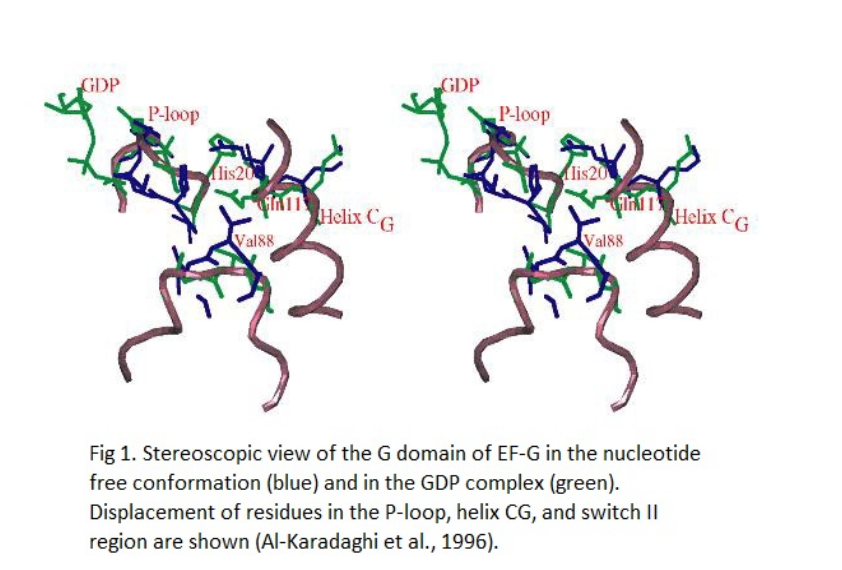
Despite this change, the P-loop maintains stabilization after GDP
incorporation. This can be attributed, in part, to the unchanged
hydrogen bond distance of His20 and
. His20 and Gln117 move positions with respect to other residues, and
this induces a displacement in the helix CG
from its position in the nucleotide free state. The CG
is then packed between helix BG and the domain
V. Both the changes in the P-loop and helix
CG affect switch II region. The switch II region is important
for the conformational changes during GTP hydrolysis to GDP
in GTPases, and this has been shown in EF-Tu (Kjeldgaard et al.,
1993). When comparing EF-G�GDP with the nucleotide free state,
however, the switch II region does not cause as much conformational
transitions as it might after GTP hydrolysis (Al-Karadaghi et al.,
1996). Nonetheless, EF-G binding of the ribosome is associated with
its own interactions, and they may contribute to the conformational
changes seen in EF-G between the pre-translocation and
post-translocation ribosome (Gao et al., 2009).
V. Ribosome Binding
EF-G is known to have different conformational states in the ribosome.
As described, the structure of EF-G is influenced by whether it is bound
to a phosphorylated nucleotide, and if so, whether it is bound to GTP or
GDP. However, the interaction with the ribosome also contributes to
conformational changes in EF-G in tandem with GTP hydrolysis. There are
two major structural changes seen in the EF-G ribosome complex: the
compact and elongated forms. The compat form is associated with GTP
binding and pre-translocation; the elongated form is associated with GDP
and post-translocation, similar to what is shown in the
three-dimensional structure to the left. Nevertheless, it remains
unclear how exactly GTP hydrolysis in EF-G is coupled with the pre- and
post-translational ribosome (Lin et al., 2015).
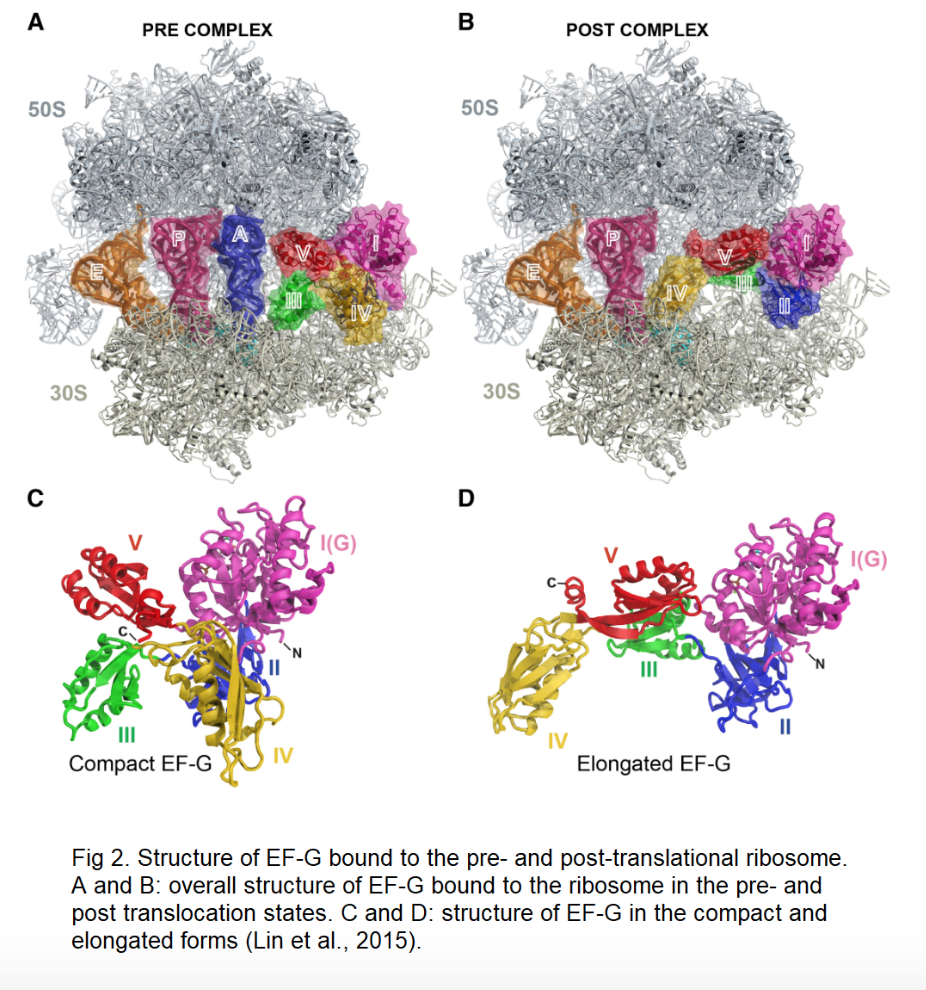
The binding of EF-G to the ribosome is similar to other GTPases,
namely, EF-Tu in complex with GTP. The major binding sites of EF-G and
the ribosome are
specific. The G domain, domain
III, and domain V surround the
sarcin-ricin loop of the ribosome. Domain III
makes
with the 30S subunit, and domain V
contacts the 50S subunit. Along with Domain III,
also contacts the 30S subunit and experiences numerous
interactions and structural changes. From the compact to the elongated
form, domain IV experiences a 90 degree
rotation into the A-site of the ribosome that mimics the tRNA moiety,
and makes substantial contacts with the ribosome, tRNA and mRNA. Domain
IV contains
that facilitate binding interaction: loop
I (residues 496 to 509) and loop II
(residues 567 to 579). Before the IV domain
occupies the A site, two
(residues 502 and 503) help flip loop
I into the P-site between the tRNA and codon. Among other interactions,
domain IV plays a major role the EF-G ribosome complex.
VI. References
Salam Al-Karadaghi, Arnthor �varsson, Anders
Liljas, Maria Garber, and Julia Zheltonosova. 1996. The structure of
elongation factor G in complex with GDP: conformational flexibility
and nucleotide exchange. Structure Volume 4, Issue 5
495-637.
J.Czworkowskil, J.Wang, T.A.Steitz1, and
P.B.Moore1. 1994. The crystal structure of elongation factor G
complexed with GDP, at 2.7 A resolution The EMBO Journal
vol.13 no.16 pp.3661-3668.
Yong-Gui Gao, Maria Selmer, Christine M.
Dunham, Albert Weixlbaumer, Ann C. Kelley, and V. Ramakrishnan.
2009. The Structure of the Ribosome with Elongation Factor G
Trapped in the Posttranslocational State. Science Vol.
326, Issue 5953, pp. 694-699.
Rajendra K. Agrawal, Pawel Penczek, Robert
A. Grassucci, and Joachim Frank. 1998. Visualization of elongation
factor G on the Escherichia coli 70S ribosome: The mechanism of
translocation PNAS 95(11)6134-6138.
A.Avarsson, E.Brazhnikov1, M.Garberl,
J.Zheltonosova Yu.Chirgadze, S.AI-Karadaghi, L.A.Svensson, and
A.Liljas. 1994. Three-dimensional structure of the ribosomal
translocase: elongation factor G from Thermus thermophilus The
EMBO Journal vol.13 no.16 pp.3669-3677.
Back to Top