PDX1 Pancreatic and
Duodental Homeobox 1
Oliver Kendall Sarah Metzmaier
Contents:
I. Introduction
The Pancreatic and Duodental Homobox 1 (PDX1) is a well
conserved homobox protein belonging to the Parahox family,
involved in the development of the pancreas and duodenum, cell
differenciation of pancreatic beta-cells, and ensuring proper
function of the mature andocrine pancreas. Homobox proteins share
a consensus sequence, in which a series of conserved amino acids
in helix 3 and the N-terminus arm
enable the sequence-dependent recongniton of dsDNA.
The final stages of pancreas development involves the production
of different endocrine cells, including insulin-producing beta-cells
and glucagon-producing alpha-cells. Pdx1 is necessary for beta-cells
maturation where developing beta-cells co-express Pdx1, NKX6-1, and
insulin. Following cell differentiation and maturation of the
pancreas, Pdx1 expression is primarily restricted to beta-cells,
while low levels of Pdx1 are expressed in alpha and sigma-cell types.
Pdx1 also maintains glucose homeostasis by regulating
transcription of insulin and other beta-cell specific genes, such as
glucokinase, islet-amyloid polypeptide (IAPP) and glucose
transporter type 2 (GLUT2) genes. By extension of the TAAT
sequence preference common to all homeobox proteins, Pdx1 binds in
a sequence specific manner to dsDNA regions containing the core
binding site (5-TAAT-3).
Mutations in the human pdx1 gene are linked to an early onset form
of non-insulin-dependent diabetes mellitus.
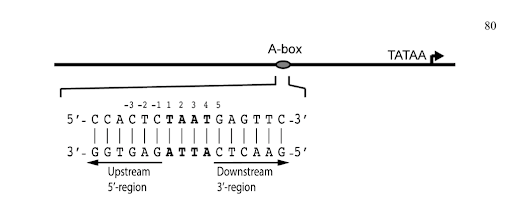 Figure 1. Depiction of Pdx1 binding elements with respect to the transcription start site. The 5’-region refers to nucleotides upstream of the core binding site and the 3’-region to bases downstream. Numbering sequence above DNA sequence is based off the first base in the 5’-TAAT-3’ sequence [1] (Bastidas 2015).
Figure 1. Depiction of Pdx1 binding elements with respect to the transcription start site. The 5’-region refers to nucleotides upstream of the core binding site and the 3’-region to bases downstream. Numbering sequence above DNA sequence is based off the first base in the 5’-TAAT-3’ sequence [1] (Bastidas 2015).
II. General Structure
Pdx1
is 283 amino acids in length; the homeodomain being 60 residues
is only about 20% of its primary sequence. The regions outside
the homeodomain have not been structurally characterized. The
non-homeodomain regions of Pdx1, which make up a large portion
of the protein, are predicted to be disordered based on the
amino acid sequence. However, there is no concrete evidence to
suggest whether the non-homeodomain regions of Pdx1 are truly
unstructured or whether they adopt residual structure.
The overall conformation of Pdx1 is similar to that of other
homodomain structures. The protein folds into three R-helices,
with helices 1 and 2
antiparallel to each other and perpendicular to helix 3, and
a flexible N-terminal arm.
Helix 3, also known as the recognition
helix,
binds in the major groove of the DNA, while the N-terminal
arm contacts the DNA bases through the minor groove.
Pdx1 also has a long disordered C-terminal tail and disordered
N-terminal domain. Partial sequence alignment of Pdx1 (homeodomain
and C- terminus) demonstrates a significant conservation of the
homeodomain, but less conservation of the disordered C-terminal
tail across species. Along with a well conserved a positively
charged patch proximal to the homeodomain. Pdx1 also interacts
with a protein subunit of a large ubiquitin ligase complex, in
which the Pdx1 C-terminus is the substrate.
III. DNA Binding
Homeobox proteins are vital DNA-binding transcription
factors that control the spatial and temporal expression of
cells and tissues. These proteins are especially important
during early cellular development. These proteins are necessary
for other biological processes, including the activation of
genes. The motif of DNA-binding homeodomain seen in homeobox
proteins bind sequence specifically to dsDNA containing the core binding site (5-TAAT-3)
.
Pdx1 shows different binding affinities for
promoter-derived (islet amyloid polypeptide (IAPP) and insulin)
and consensus-derived DNA sequences. This suggest the presense
nucleotide sequence discrimination in the flanking regions. Data
has demonstrated that Pdx1 preferentially bound sequence specific
insulin elements containing a pentameric DNA sequence (5-CTAAT-3)
rather than IAPP elements encompassing a (5-TTAAT-3) sequence. The
binding specificity of Pdx1 is largely influenced by the presence
of a TAAT sequence, which data has proven does not provide enough
information to convey specificity entirely.
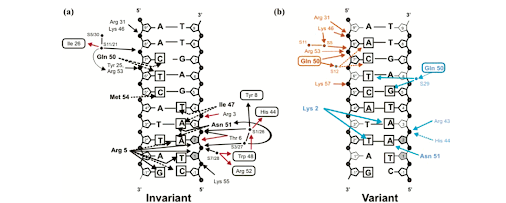 Figure 2. Diagrams showing the invariant (a) and variant (b) contacts between DNA and Pdx1. Direct Pdx1/DNA interactions as well as indirect protein/DNA interactions mediated through water molecules are represented. Sugars are represented as pentagons and phosphates as black circles; hydrogen bonds as solid lines and van der Waals contacts as dashed lines. Black arrows represent contacts by protein side chains; red arrows represent contacts by the protein backbone. Amino acid side chains contacting phosphates or sugars of the DNA are shown in normal text; amino acids contacting DNA bases are in bold; amino acids contacting DNA through a water molecule are in boxes. The DNA bases in contact with protein are marked with boxes around the base name. Sugars in contact with protein are shaded gray. (b) Residues participating in contacts specific to model A are in orange text and those specific to model B are in blue text (Longo et al. 2007).
Figure 2. Diagrams showing the invariant (a) and variant (b) contacts between DNA and Pdx1. Direct Pdx1/DNA interactions as well as indirect protein/DNA interactions mediated through water molecules are represented. Sugars are represented as pentagons and phosphates as black circles; hydrogen bonds as solid lines and van der Waals contacts as dashed lines. Black arrows represent contacts by protein side chains; red arrows represent contacts by the protein backbone. Amino acid side chains contacting phosphates or sugars of the DNA are shown in normal text; amino acids contacting DNA bases are in bold; amino acids contacting DNA through a water molecule are in boxes. The DNA bases in contact with protein are marked with boxes around the base name. Sugars in contact with protein are shaded gray. (b) Residues participating in contacts specific to model A are in orange text and those specific to model B are in blue text (Longo et al. 2007).
The Recognition Helix and the Major Groove.
Helix 3
of Pdx1 forms specific interactions in the major groove with
the bases of Ade 2, Ade 3, and Thy 4
of the TAAT core, and the bases of Cyt
5, Thy 6, and Cyt 7.
Recognition of the TAAT core in the two complexes is through
van der Waals contacts made by Ile 47
with Ade 3 and Thy 4, and
through two hydrogen bonds by Asn 51
with Ade 3. Asn 51 also forms a hydrogen bond with Ade
2. Bases Cyt 5, Thy 6, and Cyt 7 are recognized through van der
Waals contacts with Gln 50 and Met 54
.
The N-Terminal Arm and the Minor Groove.
In Pdx1, the N-terminal sequence contains three basic residues,
Lys 2, Arg 3, and the strictly conserved Arg 5 in position 5
. Arg5
interacts within the narrow minor groove of the DNA where it
hydrogen bonds to the ribose sugar and O2 of Thy1
, in addition
to12 the ribose sugar of Ade2. Arg 3 does not contact the DNA
bases, but positions the N-terminal arm by reaching into the major
groove to interact with the N? nitrogen of Arg 43 on helix 3
. In
turn, the guanidinium nitrogens of Arg 43 form hydrogen bonds with
both backbone phosphate oxygens of Ade 3
in the major groove.
Arg 43also forms a hydrogen bond with His 44
.
Lys 2 protrudes into
the minor groove to make contact with the base of Thy 2 and forms
a weak hydrogen bond with Ade 3
.
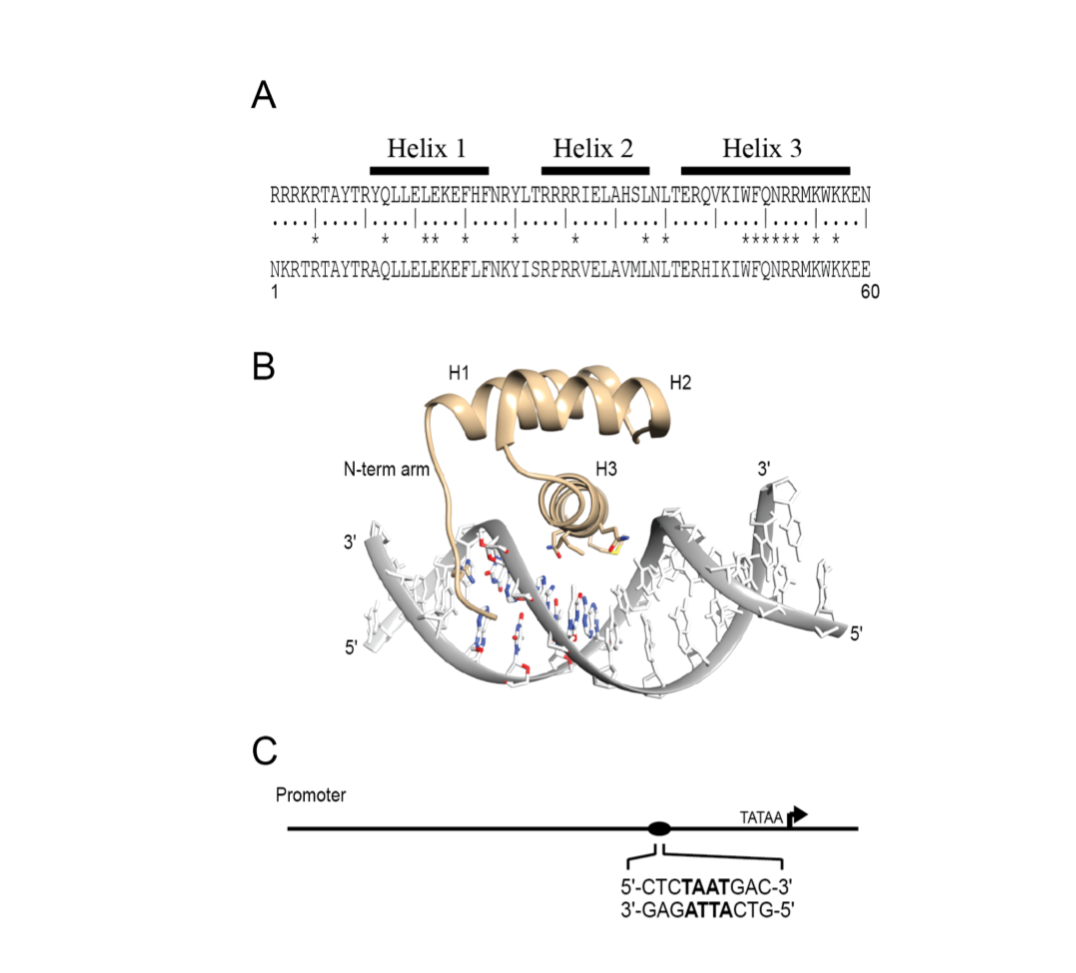 Figure 3. Canonical homeodomain sequence and structure. (A) The consensus amino acid sequence (aa 1-60) derived from 346 homeodomains (upper) and the Pdx1 homeodomain sequence (lower).1 Highly conserved amino acids are marked by asterisks. (B) Crystal structure of the DNA-bound configuration of the homeodomain motif displaying the disordered N-terminal arm and the three helices (pdb ID: 2H1K). (C) Representation of a promoter region, where the directionality of the binding element is defined relative to the transcription start site (Bastidas 2015).
Figure 3. Canonical homeodomain sequence and structure. (A) The consensus amino acid sequence (aa 1-60) derived from 346 homeodomains (upper) and the Pdx1 homeodomain sequence (lower).1 Highly conserved amino acids are marked by asterisks. (B) Crystal structure of the DNA-bound configuration of the homeodomain motif displaying the disordered N-terminal arm and the three helices (pdb ID: 2H1K). (C) Representation of a promoter region, where the directionality of the binding element is defined relative to the transcription start site (Bastidas 2015).
IV. Arg150 Binding
Arg-150
is highly conserved in all homeodomain sequences. In
homeodomain proteins, the disordered N-terminal arm inserts
into the minor groove of DNA enabling Arg-150 to interact
directly with bases.
The negative charge and narrow width of the minor
groove allows arginine and lysine to make contact within the
DNA. Arginine and lysine can therefore hydrogen bond with
bases in the minor groove, primarily to N3 in purines and O2
in pyrimidines.
Several conserved residues in the homeodomain
sequence are responsible for sequence selectivity (mostly
residues in the DNA recognition helix).
Not only is Arg-150 important for interacting with the
first two bases in the core binding site, but it is vital for
the binding of the protein. Arginine side chains carry a
positively charged guanidino group that is also capable of
bi-directional hydrogen bonding interactions. This enables
arginine to bridge two bases. Lysine side chain possess a
positively charged amino group that can play a similar role
.
A study conducted in 2015 illustrated that, Pdx1 is able to
reject A-T and G-C base pairs at the 5 prime position,
relative to the TAAT core, due to interactions between
Arg-150s and the minor groove. Interestingly, the study shows
that Pdx1 is able to discriminate between insulin and iapp
promoter-derived sequences. This discrimination is established
by the electrostatic potential created by the positioning of
their respective 5 prime flanking residues at the first T-A
pair of the core TAAT sequence.
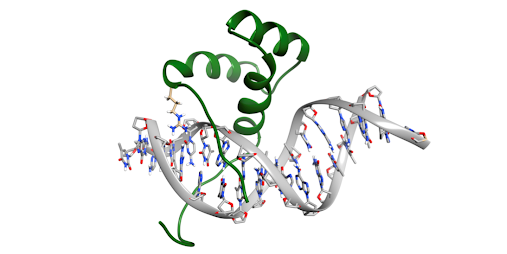 Figure 4. A representative frame from the 5 ?s molecular dynamic simulation illustrating Arg- 155 interacting in the minor groove. The side chain of Arg-155 is colored in tan, whereas the remainder of Pdx1-HDx is colored green (Bastidas 2015).
Figure 4. A representative frame from the 5 ?s molecular dynamic simulation illustrating Arg- 155 interacting in the minor groove. The side chain of Arg-155 is colored in tan, whereas the remainder of Pdx1-HDx is colored green (Bastidas 2015).
VI. References
Antonella Longo, Gerald P. Guanga, and
Robert B. Rose. 2007. Structural Basis for Induced Fit
Mechanisms in DNA Recognition by the Pdx1 Homeodomain.
Department of Molecular and Structural Biochemistry 46,
2948-2957.
Bastidas, Monique. 2015. Structural and
Functional Characterization of the Transcription Factor PDX1
and its Interactions with DNA and Proteins. The Pennsylvania
State University, The Graduate School, The Eberly College of
Science, 1-191.
Back to Top