E. coli RfaH
Paula Cancelas Calvo '22 and Christopher Ponne '22
Contents:
I. Introduction
The Escherichia coli RfaH protein is a universally
conserved transcription factor of the NusG/Spt5 family and has only
been studied in Escherichia coli. This family of proteins are the only
transcription factors that have coevolved with RNAP since the last
universal common ancestor. They also have a unique method to recruit
and elongate RNA polymerase (RNAP) for the transcription of virulence
genes. RfaH, in particular, is found to activate long operons that
encode antibiotics, capsules, toxins, and pili through inhibition of
Rho-dependent termination (Zuber, 2018).
When RNAP encounters transcriptional pausing or termination,
RfaH will work as an antiterminator to assist in elongation. RfaH
binds directly to the ops sequence located in the non-template (NT)
DNA strand of the transcription bubble. Contact with the ops
sequence is thought to trigger domain dissociation, transforming
RfaH into an open, active state where the N-terminal domain
can bind to RNAP and the C-terminal domain
changes structure. Once activated, RfaH remains bound to the
transcription elongation complex (TEC) until termination. The CTD
recruits the 30S subunit of the ribosome to the template/leader
sequence where the ribosome will then translate the RNA after
transcription. After TEC dissociation, RfaH regains the
autoinhibited state and the cycle repeats (Zuber, 2018). By
understanding this protein it may be possible to inhibit RfaH and
enable termination of viral genes found in any cell, including human
cells.
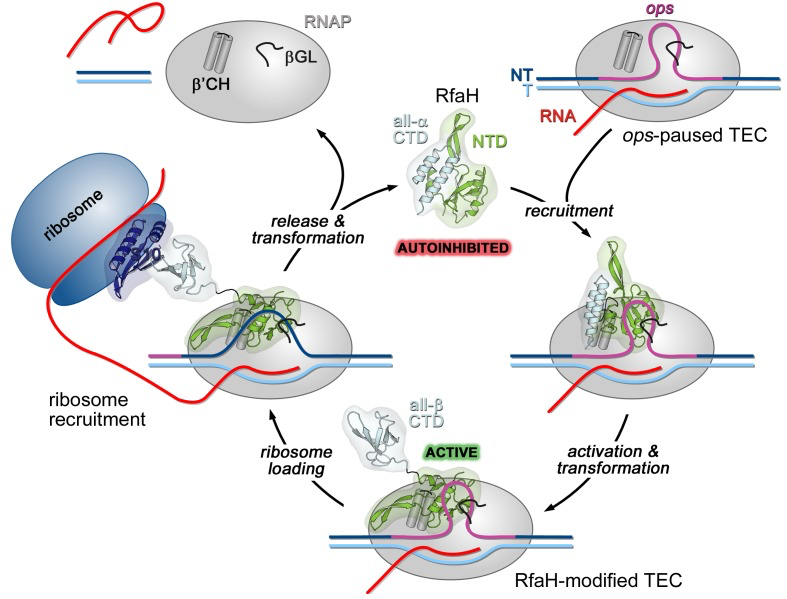
Figure 1.
RfaH recruitment to the ops-paused RNAP via the hairpin loop in the NT strand of the ops sequence. RfaH activates and upon release the CTD changes into a beta barrel. The CTD interacts with the small subunit of the ribosome to begin translation of the RNA afterwards, RfaH is finally released along with RNAP. The process repeats with another ops sequence (Zuber, 2018).
II. General Structure
Proteins of the NusG/Spt5 family usually have
N-terminal domains (NTD) of mixed alpha and beta
topology connected to beta barrel C-terminal domain
(CTD) along with a KOW motif via a flexible linker called the N/C linker.
Rfah, however, the CTD consists of two
in an alpha helical hairpin that have 2 acidic residues
at the tip of the domain (
and
). The
NTD binds across the DNA-binding channel, bridging the
RNAP beta prime clamp and beta lobe domains pincers and
resulting in RNAP becoming pause-resistant. On release,
the alpha helical CTD spontaneous forms into a beta
barrel (Zuber, 2018)
III. RfaH and ops interaction
The main difference between NusG and RfaH is that
RfaH action depends on the ops site. The ops site is
thought to play four roles during RfaH recruitment,
first it can slow down RNAP to allow more time for RfaH
recruitment. Rfah targets all have a pause-inducing TG
dinucleotide at position 11 and 12 (not shown) which
allows RNAP delay.Second, it mediates sequence-specific
binding of RfaH to the NT DNA strand exposed on the
surface of the TEC. Third, induces TEC isomerization
into a structurally distinct paused state that may be
necessary for productive recruitment of RfaH. Finally,
pausing could be required for ribosome recruitment.
T11, as previously mentioned, has
an essential role in pausing RfaH activity. Without
RfaH, RNAP pauses at C9 and U11 in the ops template
strand. With RfaH, pausing at U11 is reduced but
increases at G12. However, pausing at U11 is
dispensable for RfaH binding when RNAP transcribes
slowly (Belogurov, 2007 and Zuber, 2018). It is
important to remember that the ops sequence/site is
located in the NT hairpin.
IV. Non-template Hairpin loop interaction
with RfaH-NTD
The NT hairpin is required for RfaH
recruitment. RfaH-NTD binds to the ops sequence in
the NT DNA hairpin loop which forms when the DNA
binds to the basic patch of RfaH-NTD, opposite of
the RNAP/RfaH-CTD binding site. The
loop
consists of G4-A7 , with
flipped
out leaving the nucleobase completely exposed. The
other nucleotides of the loop make stacking
interactions.
The flipped T6 is inserted
into a deep narrow,
on Rfah-NTD, which is formed of H20, R23, Q24,
and R73 located in helices alpha 1 and alpha 2.
is
packed in the positive surface next to the
cavity T6 is located. RfaH-NTD makes contact
with nucleotides in the loop, involving K10,
H20, R23, Q24, T68, N70, A71, T72, R73, G74,and V74
.
Base specific interactions with RfaH-NTD are
made G4, G5, and T6, but only G5 and T6 form a
hydrogen bond network
and may underline sequence specific recognition.
The side chains K10,
H20, R23, and R73 directly
interact with the ops DNA. There are no aromatic
residues located near G5 and T6, therefore
contact between these two nucleotides and these
4 amino acids actually mediate specific
recognition of ops by RfaH. The hairpin stem is
formed by base pairs C3:G8 and G2:C10, with T9
being flipped out (Zuber, 2018).
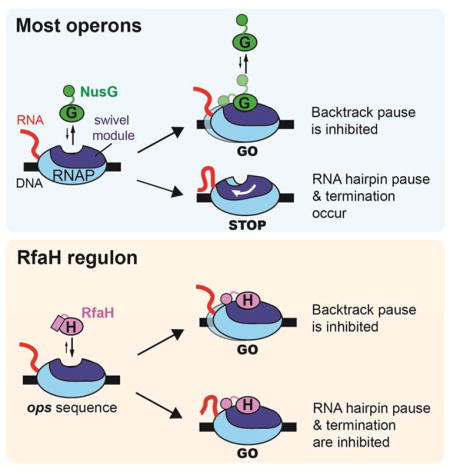
Figure
2.
VI. References
Belogurov, G.
A., Vassylyeva, M. N., Svetlov,
V., Klyuyev, S., Grishin, N.V.,
Vassylyev, D. G., and
Artsimovitch, I. “Structural basis
for converting a general
transcription factor into an
operon-specific virulence
regulator.” Molecular cell
vol. 26,1 (2007): 117-29.
Kang, J.
Y., Mooney, R. A., Nedialkov,
Y., Saba, J., Mishanina, T. V.,
Artsimovitch, I., Landick, R.,
and Darst, S. A. “Structural
Basis for Transcript Elongation
Control by NusG Family Universal
Regulators.” Cell vol.
173,7 (2018): 1650-1662.e14.
Zuber, P.
K., Artsimovitch, I.,
NandyMazumdar, M., Liu, Z.,
Nedialkov, Y., Schweimer, K.,
Rösch, P., and Knauer, S. H.
“The universally-conserved
transcription factor RfaH is
recruited to a hairpin structure
of the non-template DNA strand.”
eLife vol. 7 e36349. 9 May.
2018.
Back to Top