TAL Effector PthXo1
Rebecca Hölzel '22 and Joanna van Dyk '22
Contents:
I. Introduction
Transcription activator-like (TAL) effectors are a class of proteins
secreted by Xanthomonas bacteria and function to infect
targeted plant cells. Xanthomonas can infect a wide range of
species, but the specific TAL effector shown here, PthXo1, is secreted
by Xanothomonas oryzae and targets the Os-8N3 gene found in
rice.
TAL effectors easily enter the nucleus of the cell and bind DNA,
ultimately promoting transcription to the benefit of the bacteria. For
example, some TAL effectors target genes involved in glucose
transport. The heightened transcription provides more glucose to the
bacteria, allowing further growth and infection of the plant (Yang,
B., et al).
As TAL effectors have predictable and repetitive structures,
engineering artificial TAL effectors is feasible. A TAL effector could
be manufactured for a specific gene it would not otherwise target. It
can then act as an activator or repressor; it may also act as a
nuclease, providing another method for targeting and editing DNA.
While promising, research with TAL effectors is still in its early
stages (“TAL”).
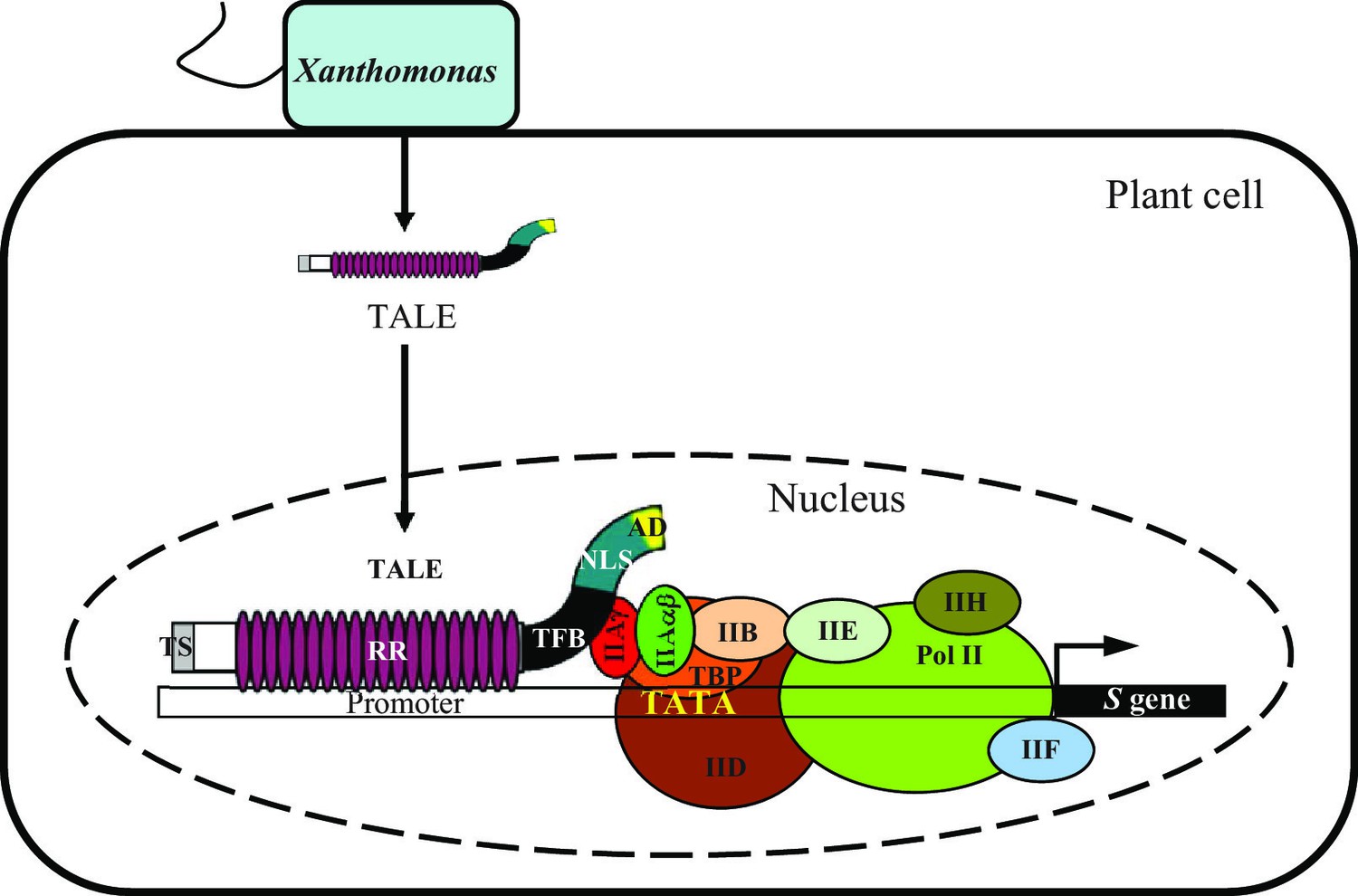
Figure 1. Xanothomonas oryzae infection of
a rice plant cell. A TAL Effector enters the nucleus and binds the
promoter region, ultimately acting in tangent with other transcription
factors to induce transcription. Image taken from Yuan, Meng, et al,
2016
II. General Structure
PthXo1 consists of a singular domain containing a repeat region as
well as the N-Terminal and C-terminal sequences. All TAL effectors,
including PthXo1, are composed of
, each composed of two alpha helices. Most repeats are 34 amino
acids in length:
Each pair of alpha helices is connected by a half-repeat loop that
contains the repeat variable diresidue (
), or amino acids in the 12th and 13th position that differ between
tandem repeats. Though more than 20 types of RVDs have been identified
in TAL effectors, the
are HD,
NG, NI,
NN, NS,
N*, and HG.
Each RVD sequence is followed by two glycine residues, though the
importance of this conserved sequence is not fully understood.
The are responsible for bacterial type III
secretion, which allows Xanothomonas oryzae to inject PthXo1
directly into the target cell's cytoplasm. The C-terminal sequences
are required for transcription activation. These sequences also
contain positively charged residues required for nuclear localization;
they act to "tag" the protein for transport into the nucleus (Mak,
A.n.s., et al).
III. Repeat Variable Diresidues and DNA Binding
The RVDs of each tandem repeat are responsible for all DNA
interactions; the remaining portion of each repeat is non-sequence
specific. Notably, each repeat, in addition to containg the RVD, also
contains a
in position 27 which creates a kink in the second helix. This kink is
essential for association of PthXo1 with the DNA, as it positions the
tandem repeat against the major groove.
Only the amino acids of RVDs contact the DNA directly. The way RVDs
bind DNA bases is predictable, which is part of the reason artificial
TAL effectors can be used to target specific DNA sequences. In PthXo1,
RVDs bind cytosine,
and
RVDs bind thymine
(one NG binds cytosine),
RVDs bind cytosine or adenine,
RVDs bind guanine, and
RVDs bind
cytosine.
Within the HD RVDs,
aspartate contacts the N4 atom of cytosine through both van der Waals
and hydrogen bonds. Glycine in both NG
and HG RVDs form non polar van der Waals interactions with the methyl group of the thymine base
The isoleucine of NI RVDs makes
van der Waals contacts with both the C8 and N7 atoms of adenine's
purine ring or C5 in cytosines pyrimidine ring.
Asparagine in NN RVDs hydrogen
bonds with the N7 in guanine or adenine; each is equally likely to
occur.
Within this structure are two N*
RVDs, where the second residue is missing. As a result, this RVD does
not contact the major groove directly. The conserved glycines
following the deleted residue are significantly further from the DNA
bases than usual, meaning N*
binding is not as specific as other for RVDs.
For most TAL effectors, the binding site is preceded by a thymine;
mutation at this position results in TAL effectors almost entirely
unable to bind the DNA. Interactions with this thymine are
accomplished by the
, where a tryptophan in position
232 makes a nonpolar van der Waals contact with thymine’s methyl carbon. (Mak,
A.n.s., et al).
V. References
Mak, A.n.s., et al. “Structure of TAL Effector PthXo1
Bound to Its DNA Target.” Science, vol. 335, Apr. 2012,
doi:10.2210/pdb3ugm/pdb.
“TAL Effectors: Function, Structure, Engineering and
Applications.”Current Opinion in Structural Biology, Feb.
2013, doi:https://doi.org/10.1016/j.sbi.2012.11.001.
Yang, B., et al. “Os8N3 Is a Host
Disease-Susceptibility Gene for Bacterial Blight of Rice.” Proceedings
of the National Academy of Sciences, vol. 103, no. 27, 2006,
pp. 10503–10508., doi:10.1073/pnas.0604088103.
Yuan, Meng, et al. “A Host Basal Transcription Factor
Is a Key Component for Infection of Rice by TALE-Carrying Bacteria.”
ELife, vol. 5, 2016, doi:10.7554/elife.19605.
Back to Top