Shown below is the 7-methyl
guanine structure within the 5' cap. Shown in
red are the methyl groups on guanine at N7 and
the ribose at positions 1 and 2. These
modifications assist in the ability of eIF4E to
recognize the 5' cap, thus promoting high
affinity binding. This leads to the recruitment
of eIF4F which signals for the initiation
complex as shown in Fig. 1 to begin translation.
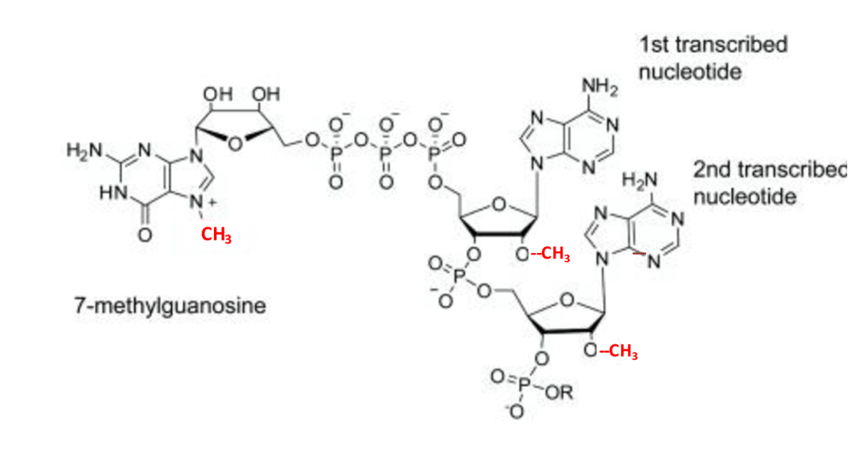
Figure 2.
Structure of the 7-methyl guanine of the
5'cap of mRNA. Methyl groups shown in red. (Cowling
et al, 2010)
IV. eIF4G and eIF4E Complex
Binding
Binding between eIF4G and eIF4E
proteins is not as well known as the
interactions between the mRNA cap and the
eIF4E protein. It is known that there is a
on the hydrophobic surface of all
known eIF4E proteins that is thought to be
important for eIF4G binding and interactions
with 4E binding proteins. Translation
initiation is thought to only proceed if
eIF4E recognizes the sequence
in eIF4G (pictured in
the button).
[Amino acid classes are labelled by
color: conserved
sequence,
hydrophobic sidechains,
polar uncharged sidechains,
electronically charged sidechains,
others.]
Marcotrigiano et al (1999)
found this sequence in mammalian eIF4GI and
eIF4GII to involve
between
hydrophobic residues Histidine-37, Valine-69,
Tyrosine-624, and Phenylalanine-628. The eIF4E
and eIF4G proteins share similar interactions
with eIF4E and the mRNA cap, including
hydrogen bonds, Van der Waals interactions,
water mediated contacts, and eIF4G also
utilizes salt bridges to contact residues. It
is also known that these peptide residues do
not hold a secondary structure unless eIF4E is
present. The mechanisms and detail about
binding between these proteins is very
limited.
V. References
Marcotrigiano, J.,
Gingras, A.-C., Sonenberg, N., & Burley,
S. (2000). Cocrystal Structure Of The
Messenger Rna 5 Cap-Binding Protein (Eif4E)
Bound To 7-Methyl-Gdp. doi:
10.2210/pdb1ej1/pdb
Marcotrigiano, J.,
Gingras, A.-C., Sonenberg, N., &
Burley, S. K. (1999). Cap-Dependent
Translation Initiation in Eukaryotes Is
Regulated by a Molecular Mimic of eIF4G.
Molecular Cell, 3(6), 707–716. doi:
10.1016/s1097-2765(01)80003-4
Rom, E., Kim, H. C.,
Gingras, A.-C., Marcotrigiano, J., Favre,
D., Olsen, H., … Sonenberg, N. (1998).
Cloning and Characterization of 4EHP, a
Novel Mammalian eIF4E-related Cap-binding
Protein. Journal of Biological Chemistry,
273(21), 13104–13109. doi:
10.1074/jbc.273.21.13104
Niedzwiecka, A.,
Marcotrigiano, J., Stepinski, J.,
Jankowska-Anyszka, M.,
Wyslouch-Cieszynska, A., Dadlez, M., …
Stolarski, R. (2002). Biophysical Studies
of eIF4E Cap-binding Protein: Recognition
of mRNA 5? Cap Structure and Synthetic
Fragments of eIF4G and 4E-BP1 Proteins.
Journal of Molecular Biology, 319(3),
615–635. doi:
10.1016/s0022-2836(02)00328-5
Cowling, V.H. (2010).
Regulation of mRNA cap methylation. The
Biochemical journal 425, 295-302.
Shanmugam, R. (2014).
Biochemical characterisation of tRNA-Asp
methyltransferase Dnmt2 and its
physiological significance.
Papadopoulos, E., et
al. “The Co-Complex Structure of the
Translation Initiation Factor eIF4E with
the Inhibitor 4EGI-1 Reveals an Allosteric
Mechanism for Dissociating eIF4G.”
Proceedings of the Natural Sciences of
America of the United States of America,
2014, doi:10.2210/pdb4tqc/pdb.
Back to Top