D. melanogaster Pumilio-Nos-Hunchback
RNA Complex
Alexandra Thoms and Erika Pontillo '23
Contents:
I. Introduction
Pumilio (Pum) and Nanos (Nos) are RNA-binding proteins that
work together as a combinatorial translational regulatory complex.
These proteins influence development, the nervous system, and the
behavior of stem cells.
In Drosophila, they are responsible for the
repression of maternal hunchback (hb) RNA. The hb
gene has multiple functions early in development, however, its
function must be repressed for some body structures to form, such
as abdominal segments. The activity of the Pum-Nos repressing
complex is sequence-specific, they recognize a particular domain
that is conserved in Pum regulated genes. Genome-wide analyses
have identified hundreds of Pu associated mRNAs, suggesting that
Pum may regulate much more than a few validated genes. The initial
recognition of RNA and binding of the complex is done by Pum,
Nanos acts as a molecular clamp that regulates and modulates the
RNA-binding and repression activities of Pumilio.
In the absence of Pum and Nos expression, the hb
protein in Drosophila is expressed throughout the embryo
and results in no abdominal segment formation. This molecular
model demonstrates how cooperative RNA binding proteins regulate
gene expression.
A
of the Pum-Nos-RNA complex demonstrates some interesting
interactions between the three components. Nos interacts with Pum
and RNA, adds to the sequence-specific contacts, and increases the
binding affinity of Pum. Nos also shifts the recognition sequence
and promotes a repression complex formation on the mRNA (Figure
1). The Pum-mRNA complex is not stably bound alone.
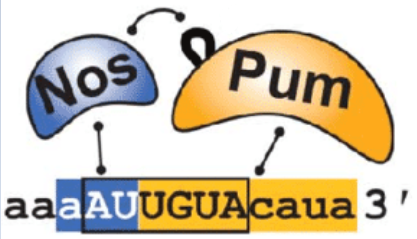
Figure 1. The Pum and Nos
RNA-binding proteins have some sequence specificity that is
adjusted when interactions between the proteins occur. (Weidmann
et al, 2016)
II. General Structure
It is important to understand how the different structures
work together on this molecular model. The amino acids side
chains on the proteins
and
bind to bases in the target
Pumilio interacts with the RNA in a
manner. Both Nanos and Pumilio also have interactions once
attached to the RNA chain. It is the
of the Nanos proteins that interact with Pumilio to repress target
gene expression.
In our example, the hunchback RNA has the
Pum-binding sequence, the binding between these two
components has a code (Section III) that can be engineered
to bind any 8 nucleotide sequence. Nanos then joins the
complex. This smaller protein increases the binding affinity
of Pum by decreasing its sequence specificity. Effector
molecules are then recruited to the complex that remove the
poly-A tail and repress translation of huncback mRNA
in the posterior abdomen of Drosophila.
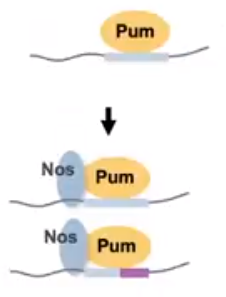
Figure 2. The
Nos protein binding to Pumilio alters the sequence
specificity of the complex while also increasing the
proteins affinity. (Hall Presentation at Kenyon
College, 2020)
III. Pumilio and RNA
Pumilio
form a curved alpha-helical domain. It can be appreciated that the
mRNA binds to the concave portion of the protein,
creating a domed binding domain on the mRNA strand.
These "PUF" protein amino acid side chain repeats
interact with the bases of the mRNA in a following a
code (Figure 3). Certain 3-amino acid sequences in the
loops connecting the alpha helices repeats bind to one
of the four bases (Table 1).
The consensus Pum-binding RNA sequence is 5'-UGUAHAUA-3' (where H can be A,U, or C).
This sequence is referred to as the
(Pumilio response element). A Pum-regulated mRNA may
have multiple PREs. Each repeat interacts with a
single base on the mRNA strand. One side chain from
the repeat interacts with one base in a
hydrogen-bonding interaction. Often another side chain
from a different Pum repeat is also involved in a
stacked interaction.
Table 1. The amino acid and
base binding involved in the Pumilio-RNA complex. The
side-chain bonding interactions are either Hydrogen
bonds or stacking. The combination of the two result
in high binding energy.
| RNA Base |
Amino Acids Involved
|
Interactions
|
View |
Uracil
(U) |
Gln, Asn,
Tyr, Asn
|
Hydrogen Bonding, Stacking, Van Der Walls
|
|
Adenine
(A) |
Gln,
Cys, Arg,
Tyr
|
Hydrogen Bonding, Stacking,
Van Der Walls
|
|
Guanine
(G) |
Glu, Ser,
Asn, Tyr
|
Hydrogen Bonding, Stacking, Van Der Walls
|
|
Cytosine
(C) |
Arg, His
|
Hydrogen Bonding, Van Der Walls
|
|
Importantly, upon binding to Nanos, the
alpha helix of Pum unfolds
to promote interactions of residues with the upstream
RNA. Specifically, Thr 1415,
Lys 1377, and Lys 1413
as they approach the phosphate groups O4 atoms on the
backbone. This is what causes the decrease in
sequence-specificity. Research shows that Nos can
stabilize binding of Pum to RNAs containing a wider
range of consensus or divergent sequences, as long as
the
sequence is not disrupted.
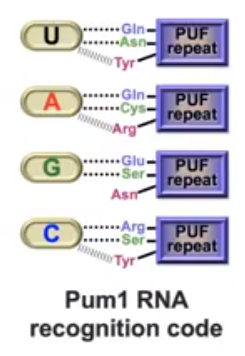
Figure 3. The
Pumillio protein of D. melanogaster and FBF
protein of C.elegans (PUF proteins) amino acid
side chains bind to the mRNA bases the same way
with each base. Van Der Walls interactions are
subjected to change based on the sequence of the
mRNA. (Wang et al, 2002)
IV. Pumilio and Nanos
Nanos and Pumilio interactions
are what drive effective translational
repression. Nos binding to hb
requires Pum-RNA recognition. The addition
of Nos at the upstream nucleotides induces
localized conformational changes in Pum that
promote Pum-Nos interactions and binding of
Pum to the RNA upstream to the core PRE.
It is
important to
the two different C-terminal regions of each
protein. The C-terminal
Pum region interacts with RNA, while the C-terminal Nos region
interacts with Pum. The C-terminal
of Pum undergoes notable changes. Loop
residues between repeats R7 and R8 rearrange
to promote Van der Waals
the of Phe 1367
with the C-terminal
alpha helix of Nos. With the deletion of its C-terminal end, Nos
cannot bind to Pum nor the target mRNA. The
mutant containing a Nos C-terminal deletion
had a strong defect in abdominal
segmentation in Drosophila,
suggesting that this interaction is
essential for in vivo activity of
the proteins (Figure 4).
Recall from the previous
section the C-terminal
the of Pum. The Pum Glu
1363 side chain is in hydrogen
bonding distance of backbone N and O atoms
of
the Ile 382.
In addition, Pum Phe
1367 forms part of a
that interacts with the Nos C-terminus Met 378.
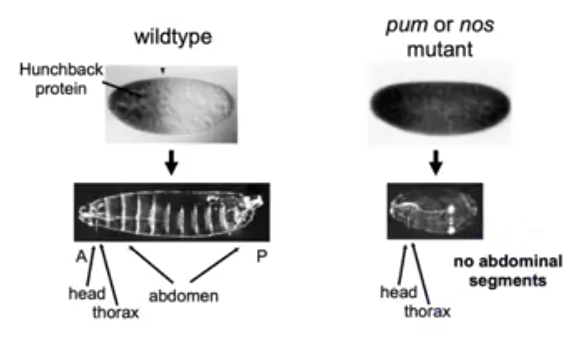
Figure 4.
The hb (hunchback) protein is constitutively
expressed in D.melanogaster when the
C-terminal end of Nos is deleted in vivo via
mutation. No abdominal segments are present in
the embryo as the Nos-Pum complex is
responsible for the gene repression in the
posterior end of the organism (weidmann et
al, 2016).
V. Nanos and RNA
Nanos binds to three nucleotides
of the PRE. The sequence is less specific
for Nos, as Nos binds based on Pumilio
recognition; however, the hb protein
Nos binding sequence is typically 5'-UAA-3'.
The
are essential for the Pum-Nos-RNA complex to
form. They
the RNA bases immediately upstream of the
PRE and, together with the C-terminal
region of Nos, embrace
the RNA and Pum. The ternary complex has a
higher affinity to the target mRNA than Pum
alone (Figure 5) Both pieces of evidence
suggests that Nanos acts as a molecular
clamp in this complex.
The first base (-1U) is inserted into a
formed by Phe 321,
Thr 366, and Tyr 369. This brings
the O4 atom of -1U into close
to the main chain N-atom of Thr
366. Similar interactions happen
with RNA bases -2A and -3A. A mutant of Lys 368 had decreased
repression activity. This is because there
is an important salt bridge
between the amino acid and the -1U phosphate
group.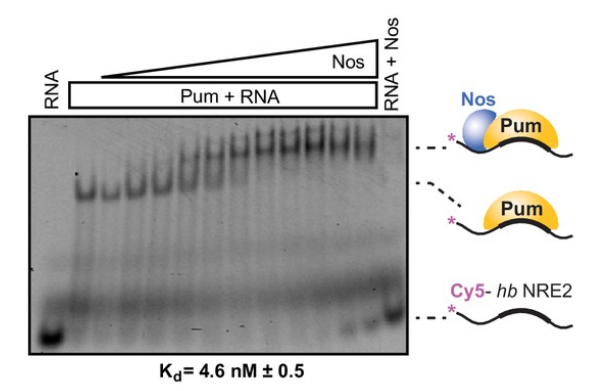
Figure 5.
A gel shift assay shows how Nos
increases Pumilio's binding affinity to
the hb RNA in D.
melanogaster.The ternary complex
formation (all three components) allows
for Nanos to increase the binding
affinity of the regulatory complex to
RNA. (Weidmann et al, 2016).
VI. References
Barker, D. D., Wang, C.,
Moore, J., Dickinson, L. K., Lehmann,
R. 1992. Pumilio is essential for
function but not for distribution of
the D. melanogaster abdominal
determinant Nanos. Genes and
Development 6:2312-2326.
Wang, C., Lechmann,
R. 1991. Nanos is the localized
posterior determinant in Drosophila. Cell 66:637-647.
Wang, X., McLachlan,
J., Zamore, P. D., Hall, T. M. 2002.
Molecular recognition of RNA by a human
pumilio-homology domain. Cell
110:501-512.
Weidmann, C., Qiu, C.,
Arvola, R., Lou, T., Killingsworth, J.,
Campbell, Z., Hall, T., Goldstrohm, A.
2016. Drosophila Nanos acts as a
molecular clamp that modulates the
RNA-binding and repression activities of
Pumilio.Biophysics and Structural
Biology | Developmental Biology and Stem
Cells.
DOI: 10.7554/eLife.17096.
Back to Top