D. melanogaster
Pumilio-Nos-hunchback RNA complex
Erika Pontillo & Alex Thoms '23
Contents:
-
I. Introduction
II. General Structure
III. Pumilio and RNA
IV. Pumilio and Nanos
V. Nanos and RNA
VI. References
I. Introduction
Pumilio (Pum) and Nanos (Nos) are RNA-binding proteins that work together as a combinatorial translational regulatory complex. These proteins influence development, the nervous system, and the behavior of stem cells.
In Drosophila, they are responsible for the repression
of maternal hunchback (hb) RNA. The hb gene has
multiple functions early in development, however, its function must be
repressed for some body structures to form, such as abdominal
segments. The activity of the Pum-Nos repressing complex is
sequence-specific, they recognize a particular domain that is
conserved in Pum regulated genes. Genome-wide analyses have identified
hundreds of Pu associated mRNAs, suggesting that Pum may regulate much
more than a few validated genes. The initial recognition of RNA and
binding of the complex is done by Pum, Nanos acts as a molecular clamp
that regulates and modulates the RNA-binding and repression activities
of Pumilio.
In the absence of Pum and Nos expression, the hb
protein in Drosophila is expressed throughout the embryo and
results in no abdominal segment formation. This molecular model
demonstrates how cooperative RNA binding proteins regulate gene
expression.
A
of the Pum-Nos-RNA complex demonstrates some interesting
interactions between the three components. Nos interacts with Pum and
RNA, adds to the sequence-specific contacts, and increases the binding
affinity of Pum. Nos also shifts the recognition sequence and promotes
a repression complex formation on the mRNA (Figure 1). The Pum-mRNA
complex is not stably bound alone.
Figure 1. The Pum and Nos RNA-binding proteins have some sequence specificity that is adjusted when interactions between the proteins occur. (Weidmann et al, 2016)
II. General Structure
It is important to understand how the different structures work
together on this molecular model. The amino acids side chains on the
proteins
and
bind to bases in the target
.
Pumilio interacts with the RNA in a manner. Both Nanos and Pumilio also have interactions once attached to the RNA chain. It is the of the Nanos proteins that interact with Pumilio to repress target gene expression.
In our example, the hunchback RNA has the Pum-binding
sequence, the binding between these two components has a code
(Section III) that can be engineered to bind any 8 nucleotide
sequence. Nanos then joins the complex. This smaller protein
increases the binding affinity of Pum by decreasing its sequence
specificity. Effector molecules are then recruited to the complex
that remove the poly-A tail and repress translation of huncback
mRNA in the posterior abdomen of Drosophila.
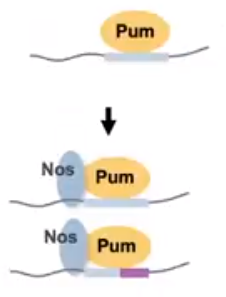
Figure 2. The Nos protein binding to Pumilio alters the sequence specificity of the complex while also increasing the proteins affinity. (Hall Presentation at Kenyon College, 2020)
III. Pumilio and RNA
Pumilio
form a curved alpha-helical domain. It can be appreciated that the
mRNA binds to the concave portion of the protein, creating a
domed binding domain on the mRNA strand. These "PUF" protein
amino acid side chain repeats interact with the bases of the
mRNA in a following a code (Figure 3). Certain 3-amino acid
sequences in the loops connecting the alpha helices repeats
bind to one of the four bases (Table 1). The consensus
Pum-binding RNA sequence is 5'-UGUAHAUA-3' (where H can be
A,U, or C). This sequence is referred to as the PRE (Pumilio
response element). A Pum-regulated mRNA may have multiple
PREs. Each repeat interacts with a single base on the mRNA
strand. One side chain from the repeat interacts with one base
in a hydrogen-bonding interaction. Often another side chain
from a different Pum repeat is also involved in a stacked
interaction.
Table 1. The amino acid and base binding involved in the Pumilio-RNA complex. The side-chain bonding interactions are either Hydrogen bonds or stacking. The combination of the two result in high binding energy.
| RNA Base | Amino Acids Involved
|
Interactions |
View |
| Uracil (U) |
Gln, Asn, Tyr, Asn |
Hydrogen Bonding, Stacking, Van Der Walls |
|
| Adenine (A) |
Gln, Cys, Arg, Tyr |
Hydrogen Bonding, Stacking, Van Der
Walls |
|
| Guanine (G) |
Glu, Ser,
Asn, Tyr |
Hydrogen Bonding, Stacking, Van Der Walls |
|
| Cytosine (C) |
Arg, His |
Hydrogen Bonding, Van Der Walls |
|
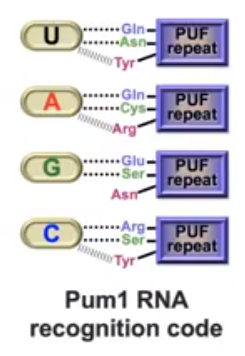
Figure 3. The Pumillio protein of D. melanogaster and FBF protein of C.elegans (PUF proteins) amino acid side chains bind to the mRNA bases the same way with each base. Van Der Walls interactions are subjected to change based on the sequence of the mRNA. (Wang et al, 2002)
IV. Pumilio and Nanos
Nanos and Pumilio interactions are what
drive effective translational repression. Nos
binding to hb requires Pum-RNA recognition.
The addition of Nos at the upstream nucleotides
induces localized conformational changes in Pum that
promote Pum-Nos interactions and binding of Pum to
the RNA upstream to the core PRE.
The C-terminal of Pum undergoes notable changes.
Loop residues between repeats R7 and R8 rearrange to
promote
of Phe 1367 with the C-terminal
alpha helix of Nos. Phe 1367 and the
C-terminal end of Nos have Van Der Waals .With the
deletion of the C-terminal end of Nos, the remaining
molecules of the protein cannot bind to Pumnor the
mRNA making. The mutant containing a Nos C-terminal
deletion had a strong defect in abdominal
segmentation of Drosophila, suggesting that
this interaction is essential for in vivo
activity of the proteins (Figure 4).
Glu 1363
and Phe 1367 abrogates
ternary complex formation, aligning the protein for
proper binding with Pum. Ile
382 and Met 378
of the Nos protein interact with Pumilio
alpha-helices through covalent bonding.
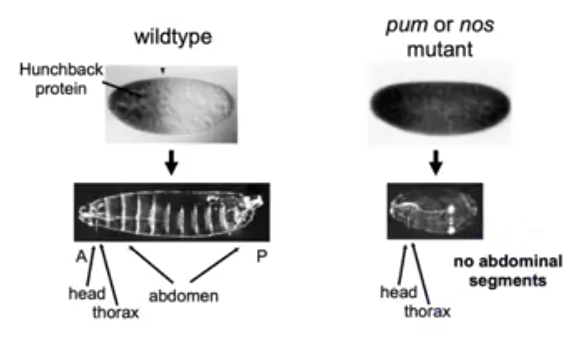
Figure 4. The hb (hunchback) protein is constitutively expressed in D.melanogaster when the C-terminal end of Nos is deleted in vivo via mutation. No abdominal segments are present in the embryo as the Nos-Pum complex is responsible for the gene repression in the posterior end of the organism (Tautz et al, 1988).
V. Nanos and RNA
Transcription activation by CAP requires more than merely the binding of cAMP and binding and bending of DNA. CAP contains an "activating region" that has been proposed to participate in direct protein-protein interactions with RNA polymerase and/or other basal transcription factors. Specifically, amino acids 156, 158, 159, and 162 have been proposed to be critical for transcription activation by CAP. These amino acids are part of a surface loop composed of residues 152-166 Researchers have concluded that the third and final step in transcription activation is this direct protein-protein contact between amino acids 156-162 of CAP, and RNA polymerase.
VI. References
Gunasekera, Angelo, Yon W. Ebright, and Richard H. Ebright. 1992. DNA Sequence Determinants for Binding of the Escherichia coli Catabolite Gene Activator Protein. The Journal of Biological Chemistry 267:14713-14720.Schultz, Steve C., George C. Shields, and Thomas A. Steitz. 1991. Crystal Structure of a CAP-DNA complex: The DNA Is Bent by 90 degrees Science 253: 1001-1007.
Vaney, Marie Christine, Gary L. Gilliland, James G. Harman, Alan Peterkofsky, and Irene T. Weber. 1989. Crystal Structure of a cAMP-Independent Form of Catabolite Gene Activator Protein with Adenosine Substituted in One of Two cAMP-Binding Sites. Biochemistry 28:4568-4574.
Weber, Irene T., Gary L. Gilliland, James G. Harman, and Alan Peterkofsky. 1987. Crystal Structure of a Cyclic AMP-independent Mutant of Catabolite Activator Protein. The Journal of Biological Chemistry 262:5630-5636.
Zhou, Yuhong, Ziaoping Zhang, and Richard H. Ebright. 1993. Identification of the activating region of catabolite gene activator protein (CAP): Isolation and characterization of mutants of CAP specifically defective in transcription activation. Proceedings of the National Academy of Sciences of the United States of America 90:6081-6085.