Receptor Binding Domain of
SARS-CoV2 Spike Protein and the ACE2 Receptor
Victoria Brown '23 and Iggy Nah '23
Contents:
I. Introduction
The SARS-CoV-2 virus (comonly referred to as the Coronavirus)
has disrupted our world for the past year. This highly contagious
virus has been diligently studied for the past several months, in
hopes of acquiring a vaccine. As of now, we know that the SARS-CoV-2
virus enters the body and interacts with the ACE2
receptor, causing several moderate to severe symptoms. Fortunately,
we now have an mRNA vaccine that encodes for the spike protein. This
vaccine will allow our immune systems to create antibodies that will
recognize the spike protein and fight off the real COVID-19 virus.
Below, we discuss the the structure of ACE2
and the receptor-binding domain (RBD)
of SARS-CoV-2 after the interaction has occured.
The Angiotensin-converting enzyme 2
receptor recognizes the
RBD of the
of the
SARS-CoV-2 virus. The glycosylated S proteins cover the surface of the
spike protein and binds to ACE2.
The spike protein is composed of two
subunits S1 and S2, which recognize the ACE2
receptor and binds the protein to the cell membrane for viral fusion.
The binding of the RBD to
ACE2 causes the spike
protein to undergo a conformational change, which leads
to the cleavage of the S1 and S2 proteins. This event is key to inducing viral
fusion into the target cell membrane. After viral RNA is released, the viral RNA
genome undergoes replication and transcription through protein
cleavage at the replicase–transcriptase complex. The viral
transcripts are then translated, and new proteins are formed.
Figure 1 depicts the process of
the interaction between SARS-CoV-2 and the ACE2,
while Figure 2 depicts the process of
viral fusion, transcription, and expression. ACE2
is commonly found in the lung cells, small intestine cells,
endothelial cells, smooth muscle cells, neurons, and glia. With that
in mind, some of the most common sympotms are fatigue, insomnia,
difficulty of breathing, and headaches. These symptoms suggest that
the translation of the viral mRNA is occuring in the aforementioned
cells, as the viral fusion (Fig
2) of SARS-CoV-2 happens in the cells that ACE2
is located.
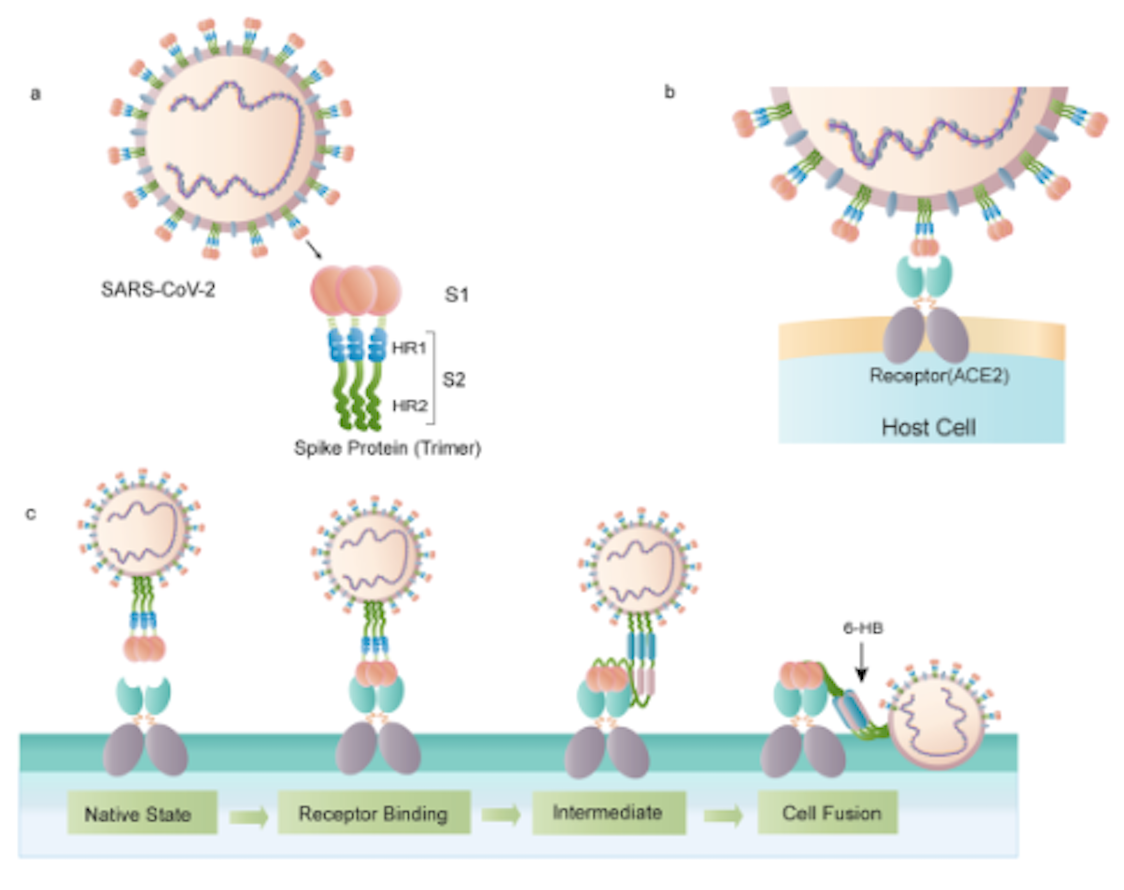 Figure 1.Interaction
between SARS-CoV-2 and the ACE2 receptor for viral fusion and
expression. (Yuan Huang, et al, 2020)
Figure 1.Interaction
between SARS-CoV-2 and the ACE2 receptor for viral fusion and
expression. (Yuan Huang, et al, 2020) 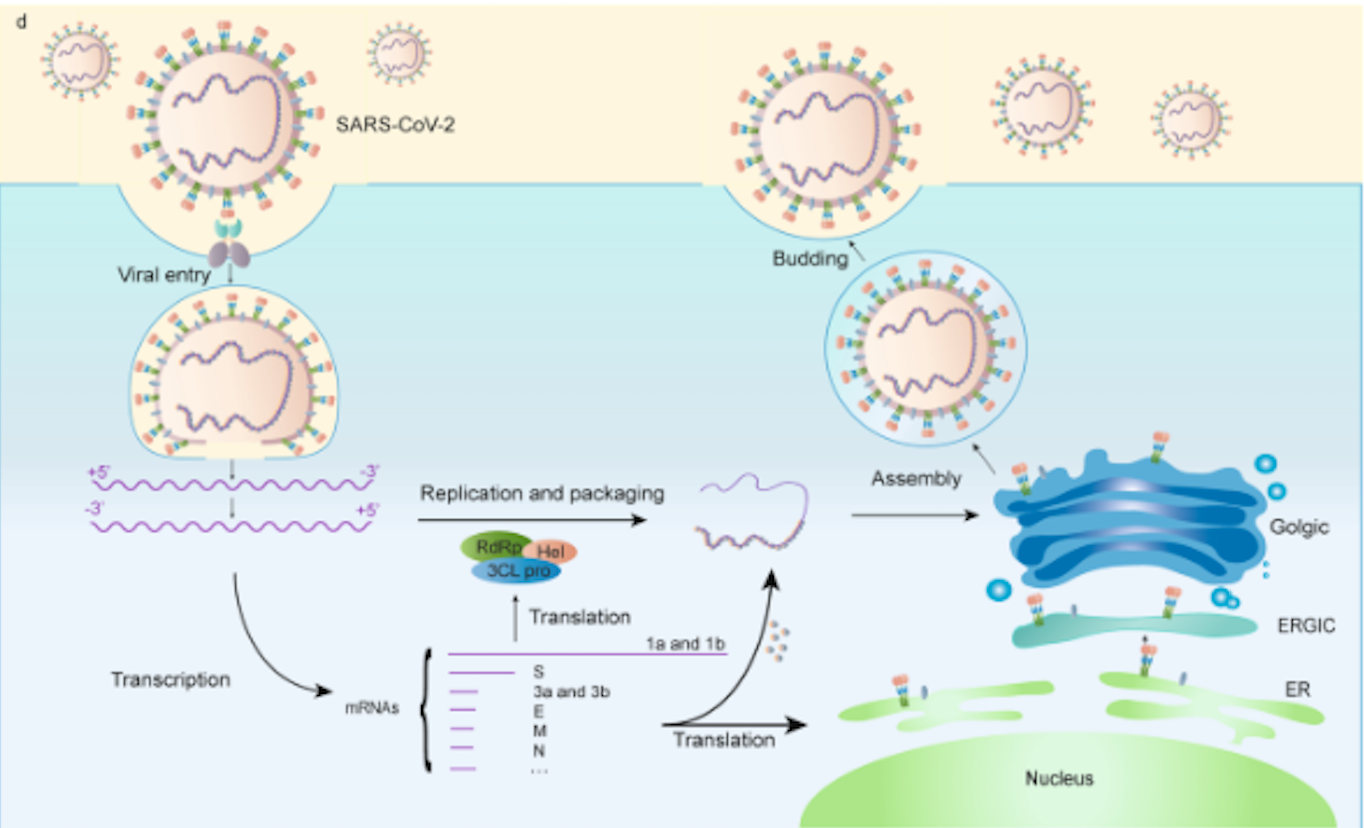 Figure 2.Fusion,
transcription, and translation of SARS-CoV-2 virus. (Yuan Huang, et
al, 2020)
Figure 2.Fusion,
transcription, and translation of SARS-CoV-2 virus. (Yuan Huang, et
al, 2020)
II. Interactions and Final Model
The precursor of viral fusion is the cleavage of the S1
and S2 subunits of the spike protein.
When the RBD binds to ACE2,
it causes S2 to change conformation and cleave from S1 as a
result.This conformational change causes the insert of a fusion
peptide into the target cell membrane. This interaction triggers
and exposes the pre-hairpin coil of the heptapeptide repeat
sequence (HR1) domain, and consequently initiates the interaction
between the HR2 domain and the HR1 trimer to form a 6-helical bundle
essential for viral fusion. This draws the cell membrane
closer for viral fusion and entry. The final model of the ACE2-RBD
complex is depicted here, in which cleavage has already occured
and part of the RBD is bound
inside of ACE2. This final model
includes the RBD , the ACE2
receptor, a
ion, a
ion,
N-acetyl-B-glucosaminide
, and 80
molecules. The spike protein S2 and SARS-CoV-2 virus are
not shown in this model.
III. General Structure
The SARS-CoV-2 spike protein
consists of an S protein on the surface of the virus that
binds their cellular receptors. The S protein consists of an
N-terminal, a transmembrane domain in the
membrane, and a short C-terminal segment.
The S protein consists of amino
acids at the N-terminus, the S1 subunit, and the S2
subunit (a conserved non- RBD
region not pictured in the molecule). However, both subunits
are responsible for receptor binding and membrane fusion. The
SARS-CoV-2 RBD also consists of
four disulfide bonds formed by nine cysteine
and an
N-terminus
of ACE2
that is responsible for binding.
ACE2 is an important
regulator and cell receptor for the SARS-CoV2 spike protein. It consists of a channel
on the top of the molecule, which includes a catalytic site
that triggers the enzymatic reaction. The channel is
contains loops, helices, a portion of
a beta sheet, and a unique helical loop
between
on
its surface. ACE2 interacts with
the SARS-CoV2 S protein, leading to the binding, fusion, and
formation of the
.
IV. Receptor Binding Domain
The receptor-binding domain ( RBD )
of the SARS-CoV-2 virus is found on the S1 subunit of the spike protein and is composed of alpha
helices and several beta
(Alternate
view of them fragmented and depicted as Beta 1-5:
,
,
,
, and
.) The RBD
region binds to the ACE2 receptor
in the region of an alanine aminopeptidase. Three disulfide
bonds
,
and
reside
in the core of the RBD and
stabilize the beta sheets. One disulfide bond,
,
resides outside the core and helps to connect the end loops in
the receptor binding
. This motif
(which is a part of the RBD
core) is partly composed of alpha
helices and is
responsible for most of the interactions between RBD
and ACE2, as it conforms to the
N-terminal helix of ACE2.
V. References
Hirano, Toshio, and Masaaki Murakami.
“COVID-19: A New Virus, but a Familiar Receptor and
Cytokine Release Syndrome.” Immunity, vol. 52, no. 5,
2020, pp. 731–733., doi:10.1016/j.immuni.2020.04.003.
Hoffmann, Markus, et al. “SARS-CoV-2
Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a
Clinically Proven Protease Inhibitor.” Cell, vol. 181, no.
2, 2020, doi:10.1016/j.cell.2020.02.052.
Huang, Yuan, et al. “Structural and
Functional Properties of SARS-CoV-2 Spike Protein: Potential
Antivirus Drug Development for COVID-19.” Acta
Pharmacologica Sinica, vol. 41, no. 9, 2020, pp. 1141–1149.,
doi:10.1038/s41401-020-0485-4.
Premkumar, Lakshmanane, et al. “The
Receptor Binding Domain of the Viral Spike Protein Is an
Immunodominant and Highly Specific Target of Antibodies in
SARS-CoV-2 Patients.” Science Immunology, vol. 5, no. 48,
2020, doi:10.1126/sciimmunol.abc8413.
Tai, W., He, L., Zhang, X. et al.
Characterization of the receptor-binding domain (RBD) of
2019 novel coronavirus: implication for development of RBD
protein as a viral attachment inhibitor and vaccine. Cell
Mol Immunol 17, 613–620
(2020).https://doi.org/10.1038/s41423-020-0400-4.
Yang, J., Petitjean, S.J.L., Koehler, M.
et al. Molecular interaction and inhibition of SARS-CoV-2
binding to the ACE2 receptor. Nat Commun 11, 4541 (2020).
https://doi.org/10.1038/s41467-020-18319-6
Back to Top