Human FOXA1 protein
Andrew Pilat '25 and Kavya Thaker '25
Contents:
I. Introduction
Forkhead box (FOX) proteins belong to an evolutionarily conserved
family of winged helix transcription factors (TF) that are expressed in
organisms from yeast to humans, and possess a highly conserved DNA-binding domain (DBD),
called the "forkhead box" domain. FOX proteins tend to bind to a FOX DNA-binding element (FBE)
to regulate the transcription of their target genes. One such protein in the FOX family, FOXA1,
is a specific regulator of the p53 gene, which is a tumor suppressor gene that plays
a key role in controlling cell division and cell death. FOXA1 binds to the upstream
promoter region of p53 and stimulates the transcription of the p53 protein.
The crystallized structure of FOXA1 binding to p53-DNA also showed FOXA1 binding to an
homotypic cluster (group of adjacent binding sites for the same TF) in the TP53 promoter
region at an additional FBE (Choi et al., 2022). This indicates that FOXA1 and the promoter region of
p53 interact with a 2:1 stoichiometry. Biological assays further confirmed that to transcriptionally
activate p53, FOXA1 has to cooperatively bind to the p53 promoter in an anti-symmetrical manner, and
that the homodimerization of FOXA1 increases the cooperative binding. Once p53 is up-regulated,
anti-proliferative activity is induced in cancer cells through a decrease in cell viability and
cell proliferation.
FOXA1 is a FOX protein of special interest as its binding affinity
shows a much stronger activation of p53 compared to FOXL2 and FOXO3.
The pattern of FOXA1 binding to p53, in a DNA-mediated homodimer with protein protein interaction,
is also distinct compared with other FOX proteins. That is of note
as this dimerization pattern coincides with the predicted FOXA1 homodimer model in a
prostate cancer cell suggesting that FOXA1 plays a biological role in cancer cells.
II. General Structure
When FOXLA1-BDB1 binds to the typical
FOX DNA-binding element 5'-RYAAAYA-3'(R=A or G, Y=C or T), the minor
groove narrows
FOXLA1-BDB2 . The molecular
weight of the entire complex is 67.33 kDa and each protein has a
sequence of 102 amino acids.
Each subunit depicted in the resolved structure is comprised of
three alpha helices and three
beta sheets.
Characteristic of the FOX proteins, FOXA1 contains a major wing.
Interestingly, dimerized FOXA1 has a noticeable "wing
1" pointing in
.
The C-terminal domain of FOXA1-DBD1
with the N-terminal domain of
FOXA1-DBD2 by a magnesium ion and
water molecules . Tyr173 and
Gln184 in both subunits bind via a
water molecule while Ser174
and Ser177 from both subunits bind via
the magnesium ion.
III. DNA Binding
Due to inconsistencies with the validation, a missing
flexible linker between helix 2 and helix 3 interferes with the
correct orientation of alpha helix 3 needed to show the proper
amino acid interactions with the resolved DNA binding domains;
therefore, the bonds are not visualized in this tutorial.
FOXA1-DBD1 binds to the major groove of the FBE1
site on p53 by the
. At the FBE1 site ,
His220
with the base of T8 and
also with A9 prime via a water
molecule. Asn216 forms
with the base of A10 prime .
Arg219 produces two
with the bases A5 and
T6. Ser217
with the phosphate backbone of T8
prime .
The Wing 1 region of p53-DNA is the
through which DNA binding by FOXA1 differs from other FOX
proteins. The
of Ser242 and of A5
forms a hydrogen bond. The side chain of
Lys240 is oriented toward the major groove of DNA,
with the bases of A2 and A3;
furthermore, it performs a water-mediated interaction with
T16 prime, forming additional contacts between FOXA1 and
p53-DNA.
When FOXA1-BDB1 first binds to the typical FBE1 site of the p53 promoter,
the minor groove narrows and creates a new binding site
for FOXA1-DBD2.
FOXA1-DBD2 is thus both
to the FBE2 site (5'-GAAAAT-3') and structure
specific as its binding affinity is increased via DNA-mediated allostery.
IV. Inconsistencies
While analyzing the provided PBD, we noticed some inconsistencies
between the resolved structure and that proposed in Choi et al.
There are many reasons why this could have happened. When we contacted
support at the Protein Data Bank, they suggested that the error likely
arose during the validation process since the structure submitted
matched the published version. Their data contained twinned fractions,
crystalline aggregates of individual crystals that are joined by
symmetric relations, which were resolved by the lab. The researchers
believe that the errors arose because twinning factors are not
considered during validation and thus led to inconsistent secondary
structures.
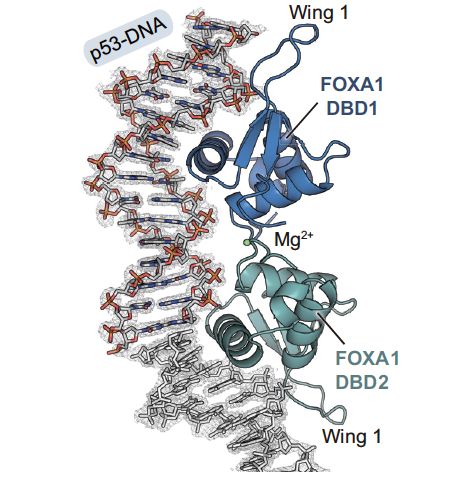
Figure 1. Schematic
Representation of FOXA1 interacting with p53 DNA. (Adapted from Choi
et al. 2022)
The deposited structure mistakenly shows
alpha helices instead of
the four imaged in Figure 1.
Pro205 through Glu209 are
supposed to be a
flexible linker that
connects the two helices that create the helix-turn-helix DNA binding
motif.
Further, the model displays three
beta sheets
instead of
found in Figure 1. This may be a consequence of the
flexible linker that was incorporated into an alpha helix. With
a lack of flexibility, a third beta sheet at
Leu191
and Thr192 confers with greater structural stability at the
tertiary level.
IV. Roles in Cancer Proliferation and Suppression
Recently, FOX proteins have emerged as critical transcriptional
regulators in cancer related processes such as tumorigenesis
and cancer progression. FOXA1 can have an oncogenic or tumor
suppressive role in human malignancies. Depending on the cellular
context, progression of the cancer, or binding interactions, either
oncogenic or tumor suppressive functionality of FOXA1 could dominate
(Parolia et al., 2019).
FOXA1 has been found to upregulate the protein levels of
p53 in colorectal cancer cells (Park et al., 2019) and low FOXA1 levels
are associated with high-grade, late-stage tumors in bladder cancer.
FOXA1 also exhibits tumor suppressive functions in breast cancer and
high expression has been found to correlate with a favorable prognosis
and improved chance of survival (Hosoda et al., 2014).
On the other hand, elevated levels of FOXA1 facilitates
prostate cancer cell growth when interacting with androgen receptors
(AR) (Robinson et al., 2014) AR-independent function of FOXA1, however,
inhibits prostate cancer metastasis (Jin et al., 2013).
Therefore, clarification of the functions of FOXA1 factors
in cancer modulation or the pathways they mediate can
potentially block metastatic progression, reverse drug
resistance when its functionality is targeted in concert
with other cancer promoting agents, or inhibit immune evasion.
VI. References
Choi,
Y., Luo, Y., Lee, S., Jin, H., Yoon, H. J., Hahn, Y., Bae, J. and Lee,
H. H. (2022). FOXL2 and FOXA1 cooperatively assemble on the TP53
promoter in alternative dimer configurations. Nucleic acids
research, 50(15), 8929 to 8946.
Hosoda, M., Yamamoto, M., Nakano, K., Hatanaka, K. C.,
Takakuwa, E., Hatanaka, Y., Matsuno, Y., and Yamashita, H. (2014).
Differential expression of progesterone receptor, FOXA1, GATA3, and
p53 between pre- and postmenopausal women with estrogen receptor-positive
breast cancer. Breast Cancer Research and Treatment, 144(2), 249 to 261.
Jin, H. J., Zhao, J. C., Ogden, I.,
Bergan, R. C., and Yu, J. (2013). Androgen Receptor-Independent
Function of FoxA1 in Prostate Cancer Metastasis FoxA1 Inhibits
Prostate Cancer Progression. Cancer research , 73(12), 3725 to 3736.
Park, Y. L., Kim, S. H., Park, S. Y., Jung,
M. W., Ha, S. Y., Choi, J. H., Myung, D. S., Cho, S. B.,
Lee, W. S., Kim, H. S., and Joo, Y. E. (2019).
Forkhead-box A1 regulates tumor cell growth and predicts
prognosis in colorectal cancer. International journal of oncology, 54(6), 2169 to 2178.
Parolia, A., Cieslik, M.,
Chu, S. C., Xiao, L., Ouchi, T., Zhang, Y., Wang, X.,
Vats, P., Cao, X., Pitchiaya, S., Su, F., Wang, R., Feng,
F. Y., Wu, Y. M., Lonigro, R. J., Robinson, D. R., and
Chinnaiyan, A. M. (2019). Distinct structural classes of
activating FOXA1 alterations in advanced prostate cancer. Nature, 571(7765), 413 to 418.
Robinson, J. L., Hickey, T. E., Warren, A. Y.,
Vowler, S. L., Carroll, T., Lamb, A. D., Papoutsoglou, N.,
Neal, D. E., Tilley, W. D., and Carroll, J. S. (2014).
Elevated levels of FOXA1 facilitate androgen receptor chromatin
binding resulting in a CRPC-like phenotype. Oncogene , 33(50), 5666 to 5674.
Back to Top