Human and Murine Nanog
Homeodomain
Logan Coleman '25 Cathy Song '24
Contents:
View Type:
I. Introduction
Nanog is an upstream transcription factor that maintains pluripotency, the ability of embryonic stem cells (ESCs) to differentiate into distinct cell types in early mammalian development. Specifically, the Nanog Homeodomain is responsible for DNA binding and sequence recognition, as well as the regulation of cell determination factors via transcription suppression. Although the exact mechanism of Nanog's gene regulation is currently unresolved, Nanog has been shown to play a critical role in gene activation and repression within ESCs. Nanog works in conjunction with a group of other transcription factors such as Oct4, FoxD3, and SOX2 that comprise an auto-regulatory loop on ESCs pluripotency (Figure 1).
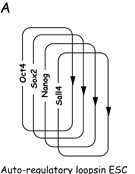

Figure 1. (A) Core transcriptional auto-regulatory loop of embryonic stem cells. Multiple transcription factors are up and down regulated to maintain a pluripotent ESC state. [6] (B) Model of transcription factor operating systems in early mouse development and ES cells. [7]
Nanog consists of three domains: the N-terminal domain, the C-terminal domain, and the Homeobox (HD) domain. Whereas the N- and C- terminal domains are generally not conserved in vertebrates and thus have not had their structures recorded, the HD domain is strongly conserved and fully resolved in multiple species. Recent interests in Nanog surround its ectopic expression in adult stem cells, restoring their proliferation and differentiation potentially normally lost due to aging or cellular senescence. [4] [7]
and
Nanog are
almost entirely structurally and functionally similar,
superimposed on top of one another with a RMSD of ~0.7 Angstrom
over the entire HD [1]. Due to limited RCSB representations of hNanog and mNanog homeodomains, either PDB will be loaded with each button to best visualize interactions. Individual monomers will be utilized for each button, but the Nanog homeodomain itself is only ever crystalized in a polymer fashion as seen in the default protein structure.
II. General Structure
The Nanog HD consists of residues 94-154 of the full-length 305 residue protein. The structure consists of three alpha-helices , with a between. H1 and H2. Helix H1 and H2 run anti-parallel , their inward folding interactions positioning H3 perpendicular. Nanog is highly diversified among vertebrates. [3] The HD is strongly conserved DNA-binding motif but Nanog HD is strongly distinct from other HD protein families. [2]
Dimerisation of the Nanog transcription factor is necessary for interactions with other pluripotency network proteins. Normally, such Dimerisation is accomplished by tryptophan repeats that are rich in the C-terminal domain. However, when two Nanog HDs are isolated, they crystallize into polymers (commonly dimers or octamers) primarily via crosslinking 2 disulfide bonds
Nanog is 8265 base pairs long, with a molecular weight of 34.6 kDa. [1]
III. DNA Binding
The region upstream of H1 is
the
, contributing to DNA binding
through minor groove contacts.
The Nanog HD contains a
which normally indicates members of the Q50 HD family, but Nanog HD
is a variant due to its relatively low sequence conservation. Nanog
instead more closely binds the sequence of the NK-2 gene family, a
central nervous system homeobox binding a C1
A2 A3 G4 X5 X6 motif. [5]
There are many noted cases of structural variants of Nanog
that alters or permits additional DNA binding. Co-variations in
affect DNA backbone contacts,
co-variations (which tend to otherwise be conserved as E17-R52 ion pairs) serve as a
mutational hotspot for structural integrity, and
, residues in the recognition
helix (highlighted), have the potential to affect affinity
and specificity of DNA binding when altered [2].
H1 and H2's
interactions position H3, the
longest helix, in a manner that promotes
of the DNA (such as the Oct4 promoter). H1 and H2 interact with the
whereas
H3 interacts primarily with the DNA backbone and H1. The prior shown
beta turn
between H1 and H2, allowing the peptide to fold into itself, biasing
interactions between the helices.
IV. OCT4 Binding
Oct4 is required for cell fate regulation in the early embryo and is down regulated upon differentiation into trophoblast cells [3]. Oct4 is also one of the most predominant factors that the Nanog dimer associates with. Nanog serves as a regulator of Oct4 transcription to maintain overall pluripotency within stem cells. [1] The primarily through H3. The resembling the NK-2 gene family is utilized in this bonding.
To test for residues responsible for Oct4 interactions, Hayashi et. al (2015) created a series of residue mutations that swapped the innate residue with Alanine. They tested bonding capabilities to Oct4 after to determine which residues were required for binding. These mutations follow the general layout of "X100A" where X was the initial '100'th residue, mutated to Alanine. Aromatic residue interactions are seen at which contact phosphates in the DNA backbone. Basic residues constitute the majority of the remaining protein-DNA interactions, seen with Other beneficial residues for Oct 4 promoter binding are seen at T100, L122, Q124, M125, Q138, T141, Q144, N145, and M148.
One key mutation, resulted in a plethora of efficacy increase. The Leucine --> Alanine showed higher affinity for OCT4 promoter DNA, greater stability of purified homeodomains, enhanced mESC self-renewal against forced differentiation by retinoic acid, and enhanced efficiency of human induced pluripotent stem cells and epiblast stem cells. The hNanog HD binding affinity increased nearly fourfold with this mutation, whereas a selection of other mutations decreased or completely abrogated DNA binding.
V. References
1. Hayashi, Y., Caboni, L., Das, D.,
Yumoto, F., Clayton, T., Deller, M. C., Nguyen, P., Farr, C. L.,
Chiu, H.-J., Miller, M. D., Elsliger, M.-A., Deacon, A. M.,
Godzik, A., Lesley, S. A., Tomoda, K., Conklin, B. R., Wilson,
I. A., Yamanaka, S., Fletterick, R. J. (2015). Structure-based
discovery of Nanog variant with enhanced properties to promote
self-renewal and reprogramming of pluripotent stem cells.
Proceedings of the National Academy of Sciences, 112(15),
46664671. https://doi.org/10.1073/pnas.1502855112
2. Chi, YI. Homeodomain revisited: a lesson from
disease-causing mutations. Hum Genet 116, 433-444
(2005). https://doi.org/10.1007/s00439-004-1252-1
3. Allouba MH, ElGuindy AM, Krishnamoorthy
N, Yacoub MH, Aguib YE. NaNog: A pluripotency homeobox (master)
molecule. Glob Cardiol Sci Pract.(2015). doi:
10.5339/gcsp.2015.36. PMID: 26566529; PMCID: PMC4625207.
4. Shahini, A., Mistriotis, P., Asmani, M.,
Zhao, R.,Andreadis, S. T. (2017). Nanog restores contractility
of mesenchymal stem cell-based senescent microtissues. Tissue
Engineering Part A, 23(11-12), 535-545.
https://doi.org/10.1089/ten.tea.2016.0494
5. Jauch, R., Ng, C. K., Saikatendu, K. S.,
Stevens, R. C.,Kolatkar, P. R. (2008). Crystal structure and DNA
binding of the homeodomain of the stem cell transcription factor
Nanog. Journal of Molecular Biology, 376(3), 758-770.
https://doi.org/10.1016/j.jmb.2007.11.091
6. Pan, G., Li, J., Zhou, Y., Zheng, H.,
Pei, D., Pan, G., Li, J., Zhou, Y., Zheng, H., Pei, D. (2006). A
negative feedback loop of transcription factors that controls
stem cell pluripotency and selfrenewal. The FASEB Journal,
20(10), 1730-1732. https://doi.org/10.1096/fj.05-5543fje
7. Shahini, A., Choudhury, D., Asmani, M.,
Zhao, R., Lei, P.,Andreadis, S. T. (2018). Nanog restores the
impaired myogenic differentiation potential of skeletal
myoblasts after multiple population doublings. Stem Cell
Research, 26, 55-66. https://doi.org/10.1016/j.scr.2017.11.018
Back to Top