Brf2: A RNA Polymerase III TFIIB Related Factor
Ashlyn Widmer '27
Contents:
View Type:
I. Introduction
TFIIB-related factor 2 (Brf2) forms a complex with the TATA binding protein (TBP) and Bdp1 (a SANT domain containing protein, directs chromatin remodeling) to recruit RNA polymerase III to promoters for Brf2 dependent genes, including those for the spliceosomal U6 snRNA and selenocysteine tRNAs. Using the human U6 snRNA promoter as a model, the Brf2 complex binds to the TATA box, an upstream region known as the proximal sequence element (PSE) is bound by SNAPc (small nuclear activating protein complex) which also binds to TBP and Brf2 before Pol III is recruited (see Figure 1).
Brf2 dependent genes contribute to essential functions within the cell such as post transcriptional regulation and tRNA production, which contribute to the growth of the cell, making their regulation crucial to the long term survival of the cell. Brf2 is redox regulated and in the presence of oxidative stress, should not be able to form the Brf2-TBP/DNA complex at the same rate in order for the cell to move to apoptosis. However, with upregulation of Brf2 and the subsequent transcription of SeCys tRNAs and production of selenoproteins, the cell is able to bypass its programmed death and proliferate rapidly, forming tumors and resulting in cancer.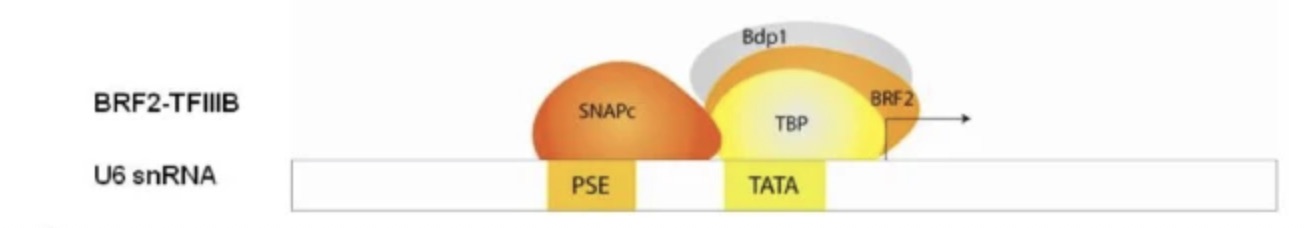
Figure 1: Illustrates the molecules involved in the formation of a transcriptionally active pre initiation complex on the Brf2 depedent human U6 promoters. From Cabarcas and Schramm, 2011.
II. General Structure
There are four main
to the Brf2 molecule: the Zn-ribbon/B-reader/B-linker (not depicted), the N-cyclin repeat, the C-cyclin repeat, and the carboxy terminal domain (CTD). The Zn-ribbon/B-reader/B-linker would typically interact with the active site of RNA polymerase III, although in this particular example, the Zn-ribbon/B-reader/B-linker was deleted prior to crystallization in order to emphasize the relationships between Brf2, TBP, and the DNA. The
interacts with the minor groove and recognizes specific bases downstream of the TATA box while the
interacts with the major groove upstream of the TATA box. The
interacts mostly with TBP and Bdp1 (not depicted). Interestingly, while TBP has beta sheets where it binds to DNA as well as some alpha helices, the Brf2
consists entirely of alpha helices and linkers.
III. Carboxy Terminal Domain
The carboxy terminal domain has three major features: the arch, TBP anchor domain, and molecular pin. The is a semi-circular α helix that is typically the site of SNAPc binding. SNAPc is an essential activator of snRNA transcription, binding to Brf2 from the PSE, but in this particular complex, the shortening of at the carboxyl terminus prevents the binding of SNAPc so RNA Pol III is not recruited and the pre initiation complex is not yet active.
The is separated from the rest of the CTD by a long linker and is the primary binding site of TBP. If the TBP anchor domain were to be deleted, not only could Brf2 and TBP not bind, but the binding of the entire complex would be compromised. In order to further strengthen the interactions between Brf2 and TBP, TBP also binds to the C-cyclin repeat through between Brf2 residues E213 and T218 and TBP residues R269 and E271.
The is found between the Brf2 C-cyclin repeat, the DNA, and TBP, essentially ‘pinning’ the entire complex together. The pin participates in via residue P359 with C-cyclin repeat residue W215 and the backbone of the TBP residue R269. The end residue of the molecular pin, , extends in between two phosphates of DNA to faciliate water mediated hydrogen bonds with the DNA at the upstream edge of the TATA box. While Brf2 and TBP can still bind normally without the molecular pin, its deletion diminishes the formation of the ternary complex as Brf2 cannot bind DNA effectively, indicating that while the N-cyclin and C-cyclin repeats are largely responsible for DNA binding, the stability of the complex relies on the molecular pin and numerous sites of Brf2-DNA interaction.
IV. DNA Binding
In addition to the stabilization offered by the molecular pin, the N-cyclin repeat contacts DNA via the minor groove downstream of the TATA box while the C-cyclin repeat interacts with DNA via the major groove upstream of the TATA box. Notably, the TBP interactions with DNA are the same regardless of whether TBP is bound to Brf2 or TFIIB.
The N-cyclin repeat contacts the DNA through a motif, where the dyad residues interact with the negatively charged phosphate backbone of the DNA (K113 with the non template strand, K114 with the template strand) to allow a short helix to enter the minor groove. This allows Brf2 to bases C+2 and T'+5 and subsequently form bonds. The side chain of residue R110 forms a hydrogen bond with base A+3 and a water mediated hydrogen bond with G'+4' , while the carbonyl oxygen of A108 interacts with G’+4 via a direct hydrogen bond. All of these cause a local distortion of the DNA, pulling the base T’+3 out of the base hydrophobic stack.
In contrast, the C-cyclin repeat contacts the DNA less intimately, with no part entering the DNA helix. Instead, the Brf2 residue forms a hydrogen bond with the nucleotide C-4. This tyrosine residue also participates in T-shaped pi-stacking with both C-4 and C-3, where the tyrosine aromatic ring is perpendicular to the aromatic rings of the two cytosine bases. This arrangement allows for attractive electrostatic interactions between the tyrosine and cytosines, further stabilizing the connection between Brf2 and DNA.
V. Redox Regulation and Relevance
Redox signaling is essential for the maintenance of cellular homeostasis. Reactive oxygen species (ROS) and reactive nitrogen species (RNS) are formed in response to irradiation or chemical damage and lead to DNA and protein damage. Due to the threats posed by ROS and RNS, cells have evolved sensing mechanisms and the ability to eliminate these reactive species. Specifically, Brf2 has two reactive cysteine residues, , that will oxidize and reduce depending on the levels of ROS in the cell. In the presence of ROS, C361 and C370 will be oxidized to form cysteine thiols, which are stabilized by the positively charged surrounding residues L363, K367, and R368.
However, only C361 affects the formation of the Brf2-TBP/DNA complex, likely due to its position on the tip of the essential molecular pin, where it directly interacts with the DNA backbone. When C361 is oxidized, DNA and Brf2 binding is reduced by a factor of fifty, which is reversed when C361 is reduced and returns to its standard state, proving that Brf2 is a redox sensor and redox regulated. One of the products of Brf2 dependent transcription is selenocysteine tRNAs, which form selenoproteins that are involved in the detoxification of ROS. When Brf2-DNA binding is inhibited, selenoproteins are no longer produced, driving the cell to apoptosis.
Many types of cancer (most prominently breast and lung cancers) have been shown to have overexpressed Brf2, indicating that when Brf2 is not inhibited by the reactive C361, the subsequent overproduction of selenoproteins allows the cell to evade apoptosis and continue growing uncontrollably. Brf2 overexpression has also been tied to patient outcomes. There is a strong association between Brf2 upregulation and poor survival rates, indicating that the more overexpressed Brf2 is, the less likely a patient is to survive their cancer. Brf2 overregulation has also been tied to metastasis risk, indicating that Brf2 may be able to be used as a biomarker for patients at risk of metastasis and a potential target for future treatments.
VI. References
Cabarcas, S.; Schramm, L. RNA Polymerase III Transcription in Cancer: The BRF2 Connection. Molecular Cancer 2011, 10 (1), 47. https://doi.org/10.1186/1476-4598-10-47.
Gouge, J.; Satia, K.; Guthertz, N.; Widya, M.; Thompson, A. J.; Cousin, P.; Dergai, O.; Hernandez, N.; Vannini, A. Redox Signaling by the RNA Polymerase III TFIIB-Related Factor Brf2. Cell 2015, 163 (6), 1375–1387. https://doi.org/10.1016/j.cell.2015.11.005.
Rashidieh, B.; Molakarimi, M.; Mohseni, A.; Tria, S. M.; Truong, H.; Srihari, S.; Adams, R. C.; Jones, M.; Duijf, P. H. G.; Kalimutho, M.; Khanna, K. K. Targeting BRF2 in Cancer Using Repurposed Drugs. Cancers 2021, 13 (15), 3778. https://doi.org/10.3390/cancers13153778.
Trachootham, D.; Lu, W.; Ogasawara, M. A.; Valle, N. R.-D.; Huang, P. Redox Regulation of Cell Survival. Antioxidants & Redox Signaling 2008, 10 (8), 1343–1374. https://doi.org/10.1089/ars.2007.1957.
Yeganeh, M.; Hernandez, N. RNA Polymerase III Transcription as a Disease Factor. Genes & Development 2020, 34 (13-14), 865–882. https://doi.org/10.1101/gad.333989.119.
Back to Top