Cytochrome P450 2B4 Enzyme
Juliette Tomamichel '26 and Juliette Levy '26
Contents:
I. Introduction
Cytochrome p450 (CYP) proteins are a class of enzymes that have a central role in cellular metabolism, notably drug metabolism. CYP enzymes become transcriptionally activated by receptor-mediated binding, and detoxify foreign or endogenous compounds. CYP enzymes are ubiquitous in cells, and belong to the monooxygenase superfamily. Due to the iron-containing , these enzymes are responsible for catalyzing oxidation reactions in the liver, intestines, and kidneys.
The cytochrome p450 2B4 (CYP2B4) enzyme belongs to the 2B subfamily, and is a heavily researched mammalian enzyme. Since CYP2B4 is a highly resolved model, it is possible to better define drug-enzyme interactions.
II. General Structure
The CYP2B4 enzyme consists of one polypeptide chain, primarily made up of alpha helices, and fewer beta sheets.
The starting
can be traced to the final
amino acid. The
group is a non-protein ring-like structure that contains a central iron (Fe) atom. The iron atom is bound to
. When bound by a substrate, the molecule is reduced from the ferric (Fe+3) to the ferrous (Fe+2) form. Once reduced to the ferrous state, oxygen is able to bind, where the oxyferrous protein accepts a second electron from electron donors (such as cytochrome P450 reductase or cytochrome b5).
CYP2B4 is made up of
. From the N terminus to C terminus: A’, A, B, B’, C, D, E, F, F’, G’, G, H, I, J, J’, K, K’, K’’, L, L’. The defining structural feature of CYP2B4 is its
opening from the surface of the protein to the heme group at the center. The cleft is
wide, and is composed of helices on both sides. Helices D, E, F, F’, G’, G, H, I, J, J’, K, K’’, L, and L' are found on
, and by helices A, A', B, B', C, and K' on the
of the cleft.
III. Open, Closed & Intermediate Conformations
CYP2B4 – one of the most flexible cytochrome p450 enzymes – adopts three conformational states: open, intermediate, and closed. For enzymatic activity, a substrate must bind to CYP2B4 while it is in the open conformation. The open conformation is formed by the large cleft, which is the active site of the enzyme. The cleft is a result of the
of residues in helix B’ from residues in helix G. As a consequence, helices F′ and G′
from the heme group.
between helices help quantify the size of the cleft in the open conformation. The three distances all average above 20 Å in length. Additionally, helix C and helix G move in a coordinated manner, where the
between the N terminus of helix C and helix G are retained, preserving the open conformation.
CYP2B4 adopts a closed conformation in the absence of a bound substrate. Without the large cleft, the
between the helices decreases. Compared to 20 Å in the open conformation, the distances are less than 15 Å the closed conformation. As CYP2B4 shifts from a closed to open conformation, an intermediate structure arises. The intermediate conformation exhibits cleft dimensions similar to that of closed and open conformations. The
between the A’ and F’ helix resembles the closed conformation more closely (13.84 Å). Although not the same, the
between the B’ and G helix is most similar to the open conformation (25.8 Å). The distance between the B’ helix and C-terminal loop is highest in the intermediate conformation (31.0 Å), in comparison to the closed (14.38 Å) and open (23.47 Å) conformation. Although current studies have not yet crystallized CYP2B4 in the intermediate structure, it is suspected that the G and B’ helix are primarily responsible for the enzyme’s conformational changes.
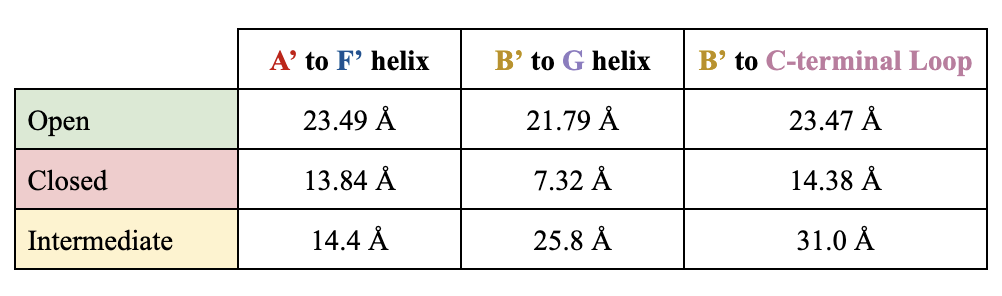
Table 1. Distances in Conformational States.
IV. Dimerization
In solution, two CYP2B4 molecules crystallize to form a
. These two molecules are found in the open conformation, where one molecule's cleft is partially filled by the F' and G' helices of the other molecule.
spanning across the F' and G' helices, are responsible for forming
that contribute to the forces holding the two monomers together. The residues in this region are all hydrophobic: serine, glutamine, valine, phenylalanine, glutamate, leucine, proline, lysine, histidine, glycine and threonine. The dimer is further stabilized by a
between His 226 and the heme iron. This covalent bond is found in the
. Ultimately, the hydrophobic interactions between the two monomers trap the dimer in the open conformation.
When CYP2B4 is not dimerized, the open conformation is not energetically or structurally stable. For this reason, in a cellular environment, the helices of CYP2B4 are better maintained in a membrane than in water. CYP2B4 can still dimerize when in the intermediate conformation, but dissociates into monomeric subunits when in the closed conformation.
V. Application
Understanding the structural functions and binding patterns of the Cytochrome P450 2B4 allows researchers to predict its interactions with a variety of drugs. The flexibility of CYP2B4’s open and closed conformational states enables diverse substrate binding. Since its conformation is only stabilized when a substrate is positioned in the active site, regiospecific and stereospecific metabolism is enabled. Dimerization is necessary for conformational changes as the CYP2B4 monomer's open structure is unstable, and favors the intermediate conformation in vivo. Ultimately, better characterization of its enzymatic activity can be manipulated for therapeutic and pharmacological purposes.
Further research is required to determine how the intermediate structure responds to substrate binding. This would allow for potential improvements in drug design and enzyme allosteric modulation. Exogenous drug metabolism could be optimized to reduce adverse reactions and boost therapeutic benefits.
VI. References
Bridges, A; Gruenke, L; Chang, Y; Vakser, I; Loew, G; Waskell, L; (1998). Identification of the Binding Site on Cytochrome P450 2B4 for Cytochrome b5 and Cytochrome P450 Reductase. Journal of Biological Chemistry, Vol 273 (27) 17036-17049 DOI: https://www.sciencedirect.com/science/article/pii/S0021925818806421.
Im, S-C; Waskell, L; (2010). The Interaction of Microsomal Cytochrome P450 2B4 with its Redox Partners, Cytochrome P450 Reductase and Cytochrome b5. Archives of Biochemistry and Biophysics, Vol 507 (1) 114-153 DOI: https://pmc.ncbi.nlm.nih.gov/articles/PMC3073529/
Li, J; Zhou, Y; Li, W; Tu, Y; (2020). Dissecting the Structural Plasticity and Dynamics of Cytochrome P450 2b4 by Molecular Dynamics Simulations. Journal of Chemical Information and Modeling, Vol 60 (10) 4417-5282 DOI: https://pubs.acs.org/doi/10.1021/acs.jcim.0c00482
Scott, E; He, Y; Wester, M; Stout D; (2003). An open conformation of mammalian cytochrome P450 2B4 at 1.6-Å resolution. PNAS, Vol 100 (23) 13196-13201 DOI: https://doi.org/10.1073/pnas.2133986100
Shah, M; Kufareva, I; Pascual, J; Zhang, Q; Stout, D; Halpert, J; (2013). A Structural Snapshot of CYP2B4 in Complex with Paroxetine Provides Insights into Ligand Binding and Clusters of Conformational States. Journal of Pharmacology and Experimental Therapeutics, Vol 346 (1) 113-120 DOI: https://pmc.ncbi.nlm.nih.gov/articles/PMC3684837/
Back to Top