Biology Dept Kenyon College |
Gene Product Interaction and Recombination |

|
Biology Dept Kenyon College |
Gene Product Interaction and Recombination |

|
|
Gene
product interaction
In real
living organisms,
all traits result from the interaction of many gene
products.
Suppose two enzymes are required to produce a trait. What may be
the result? What will happen to Mendelian inheritance ratios?
1. In sweet peas, purple is dominant; white is recessive. Suppose two recessive white flowers from different breeders are cross-bred together. The flowers--surprise--show the dominant purple color. When self-crossed, the ratio is 9 purple: 7 white. Explain these results. 2.
In fancy fowl, feathers are dominant to non-feathered. Two
pure-breeding
feathered birds from two different fowl fanciers are bred, producing
lovely
feathered offspring. The offspring are intercrossed, and a few of
their offspring have no feathers. The ratio is:
3.
Beetle colors. Suppose the color of beetles is determined by this enzyme pathway. Enzymes A and B are needed in series, but enzyme C represses enzyme A. 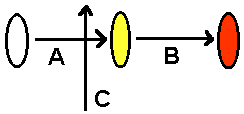 Figure out
what dihybrid
crosses would produce what ratios. Remember that you have to
specify
the entire genotype, including the purebreeding locus; for instance:
Complementation Complementation refers to the fact that mutations producing null alleles in different genes can be complemented, or compensated in effect, by the gene product provided elsewhere. An example of complementation is two-way recessive epistasis: The two white strains of sweet pea complement each other: P1 P1 p2 p2 X p1 p1 P2 P2 --> P1 p1 P2 p2 (purple flowers) You can use complementation analysis to intercross a large number of independently isolated null phenotypes, and figure out how many different genes are required for the enzyme pathway to produce the phenotype. See the MIT site on Complementation. Recombination by Crossover of Chromosomes Two DNA helices may "cross over" or recombine, a process requiring breakage of each strand at the phosphodiester backbone and ligation to another strand. There are two different classes of recombination:
Problem: Is hin recombinase involved in homologous recombination, or site-specific recombination? Explain your answer. Homologous recombination This image series shows how two homologous chromosomes within a tetrad cross over and exchange portions of their arms. (Problem: What phase of meiosis?) The blue and red arms designate homologues from different parents. (Why are they each double already?)Click the image: 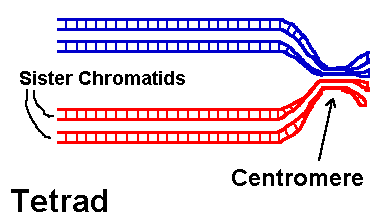
Linkage and Mapping What happens to Mendelian
ratios
when two genes are linked on the
same
chromosome?
A B
a
b
A
b
a B
A B
A
b
Test
Cross
45% A
B
5%
5%
A b
45%
5%
a
B 45%
Which classes are "parental"? "Recombinant"? Map
Distances.
Problem: What happens when there are double crossovers? How does this interfere with map calculations? LOD scores
and pedigrees.
|