Biology Dept Kenyon College |
Bacterial Gene Regulation |
 |
Biology Dept Kenyon College |
Bacterial Gene Regulation |
 |
|
Complementation Analysis Lac Operon Quiz -- Highly Recommended Organization of Genes Control of Gene Expression Bacterial Operons Analysis of Operons Promoter Structure Environmental Regulons Research Complementation Analysis Complementation means that two different sources of genetic information (usually, different gene loci encoding proteins or RNAs of different function) together each provide something the other lacks. Example: Two white-flowered plants cross to
produce purple flowers,
although purple is dominant.
Complementation analysis is easiest to do in bacteria, fungi, or C. elegans, where many mutants of a given phenotype can be obtained. If we isolate a large number of strains with the same defective phenotype,we can cross them in all combinations, and figure out the number of complementation groups. Any two defective strains that FAIL to complement are in the same complementation group. Usually each complementation group represents one of the essential enzymes in the pathway. Problem 1: Figure out how many complementation groups there are in these examples. The concept of complementation is extremely important in molecular biology. For example, the sickle-cell mouse line could only be created because two strains with different defects (lack of mouse or human globin genes) could be mated to complement each other's defects. The fact that genes from different species can complement each other was one of the most significant conceptual advances in molecular biology. Complementation is now used routinely to answer more subtle questions of how genes are regulated. You absolutely need to understand complementation to understand molecular biology. Bacterial complementation.
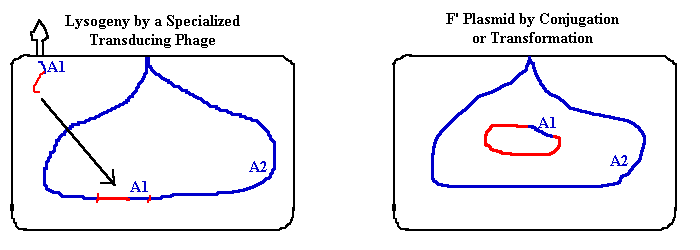 1. Specialized Transduction. A lysogenic bacteriophage can excise itself so as to carry a piece of host DNA by mistake. The phage will now carry a second copy of an allele (or linked alleles) into a host cell. The new bacterium is a partial diploid for the allele(s). In biotechnology, a phage chromosome can have a piece of foreign DNA ligated into it in the test tube. Then the phage DNA is packaged into phage, and it can infect a new host where it either (1) produces many copies of the host gene; or (2) lysogenizes the host, to express the cloned DNA. 2. F' plasmid.The F plasmid can recombine itself into the host chromosome, then recombine itself out again with some host DNA by mistake. When it enters the next host cell, it carries a second copy of several genes; again, a partial diploid is created. In biotechnology, a plasmid can have a piece of foreign DNA ligated into it in the test tube; then the plasmid is transformed into E. coli. Then the plasmid makes many copies, including the cloned gene. Suppressor mutation analysis.
Organization of Genes In bacteria, a number of gene ORFs can be organized into an operon. All the gene sequences in a given operon are transcribed on a single mRNA, starting at one promoter. An example of an operon is shown, tuf-s10, from Borrelia burgdorferi, which causes Lyme Disease: 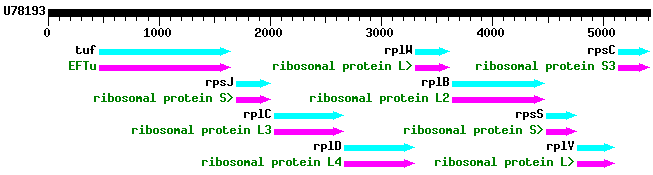 This operon encodes ribosomal proteins, and an elongation factor; in all:
Human Growth Hormone Receptor 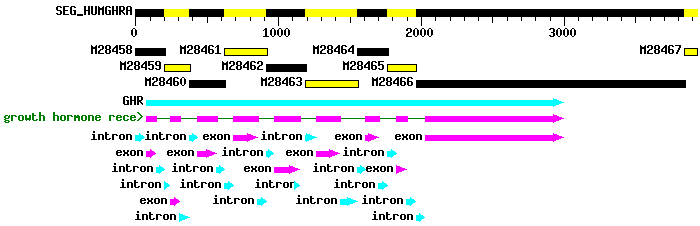 For other interesting operons, try searching GenBank, the international repository for all known DNA sequences. (Funded by the U.S. government--your tax dollars at work.) Control of Gene Expression
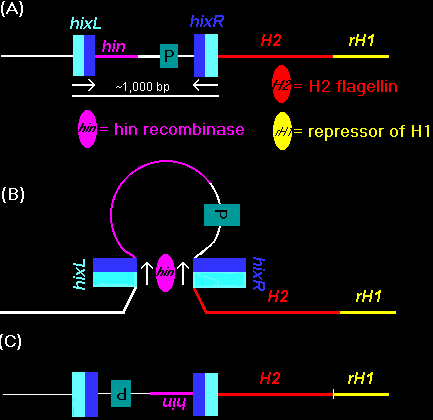 transcription is followed by translation of the H2 flagellin protein and of the rH1 protein, a repressor of H1 flagellin genetranscription.When the hin recombinase inverts the DNA segment containing the promoter, it turns off H2 transcription and de-represses H1. Transcription level regulation Regulation of transcription can respond more quickly, and is more reversible. Regulation at the level of translation is even more reversible. In BIOL 14, we will focus on regulation of transcription, or operon control. The Lac Operon
The following explanation of the Lac operon is modified from MIT
Lac Operon.
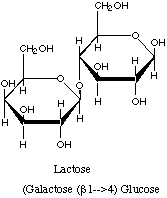 It should turn on the enzymes that are required for lactose degradation. These enzymes are:
A bacterium's prime source of food is glucose, since it does not have to be modified to enter the repiratory pathway. So if both glucose and lactose are around, the bacterium wants to turn off lactose metabolism in favour of glucose metabolism. There are regulatory sites upstream of the Lac genes that respond to glucose concentration. An overall picture of Lac regulation would be this:
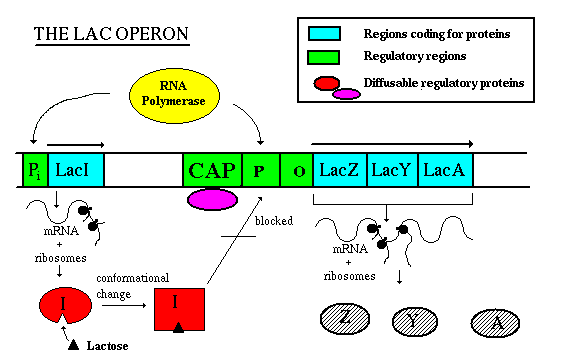
When lactose is present, it acts as an inducer of the operon. It enters the cell, rearranges slightly to form allolactose, then binds to the Lac repressor. A conformational change causes the repressor to fall off the DNA. Now the RNA polymerase is free to move along the DNA, and RNA can be made from the three structural genes. The mRNA will be translated to the proteins which transport and metabolize lactose. When the inducer (lactose) is removed, the repressor returns to its original conformation and binds to the DNA, so that RNA polymerase can no longer get past the promoter. No RNA and no protein is made. Note that RNA polymerase can still bind to the promoter though it is unable to move past it. That means that when the cell is ready to use the operon, RNA polymerase is already there and waiting to begin transcription; the promoter doesn't have to wait for the holoenzyme to bind. 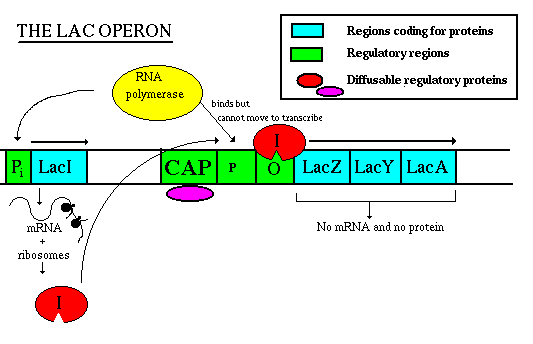 When levels of glucose (a catabolite) in the cell are high, a molecule called cyclic AMP is inhibited from forming. But when glucose levels drop, ATP phosphates are released until at last forming cAMP: ATP --> ADP + Pi --> AMP + Pi --> cAMP cAMP binds to a protein called CAP (catabolite activator protein), which is then activated to bind to the CAP binding site. This activates transcription, perhaps by increasing the affinity of the site for RNA polymerase. This phenomenon is called catabolite repression, a misnomer since it involves an activator protein, but understandable since it seemed that the presence of glucose repressed all the other sugar metabolism operons. This image shows a "close-up" view of CAP regulation: 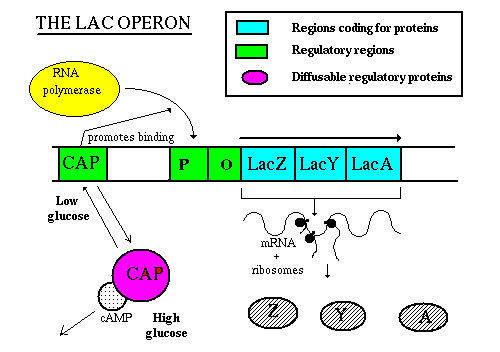 Corepressor control
Analysis of operon control What experiments do we perform to figure out how operons are regulated? We use partial diploid strains created by F' or specialized transduction. In either case, we test what happens when a strain is diploid for regulatory elements. Regulatory mutants can have various kinds of mutant phenotypes. For example: p- Promoter fails to bind
RNA polymerase. No transcription occurs.
What will happen? What kinds of complementation can occur?Does
is matter if the two mutant alleles are adjacent on the same chromosome
(cis) or separated (trans)?
Problem 2. Predict whether the following diploids produce B-galactosidase, in the presence of lactose; in the absence of lactose. Explain why.Explain in each case whether it matters if the two mutant alleles are located in cis or in trans. p + lacZ -
lacI +
p + lacZ -
lacI -
p + lacZ -
lacI +
p + o-c
lacZ + lacI + (A
constitutive operator NEVER binds repressor,
Problem 3. Explain two different genetic processes in bacteria that can create a "partial diploid" for a small part of the genome. Explain why these processes are useful for bacterial genetic analysis. Problem 4. State
whether B-galactosidase is expressed by each lac operon diploid,
(1) and (2), and briefly state why (one sentence). Complete possible
genotypes for (3) and (4).
MIT Bacterial Genetics Problems Quiz on Lac Operon
-- Highly Recommended
Molecular Structure of Promoters Promoters are defined by sequences of base pairs upstream of the transcription start site. The RNA polymerase tends to recognize promoter sequences in which most of the base pairs match the promoter consensus sequence. The consensus sequence is a composite defined by the most common base to occur at each position. Base-substitution mutations can decrease or increase the efficiency of the promoter. 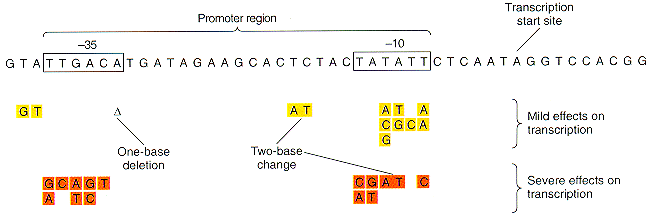 Bacterial consensus promoters include two regions of six base pairs each, at -10 and -35 bases upstream. However, no two promoters are exactly alike, and no promoter exactly matches the consensus sequence. Additional sites for environmental regulators can be found as far as -50 to -300 bases upstream. Griffiths et al, Genetic Analysis
Environmental Regulons Genes can be regulated together even though they are located at different parts of the bacterial chromosome. A group of genes regulated by the same environmental signal is a regulon. An example is the rpoS starvation regulon. 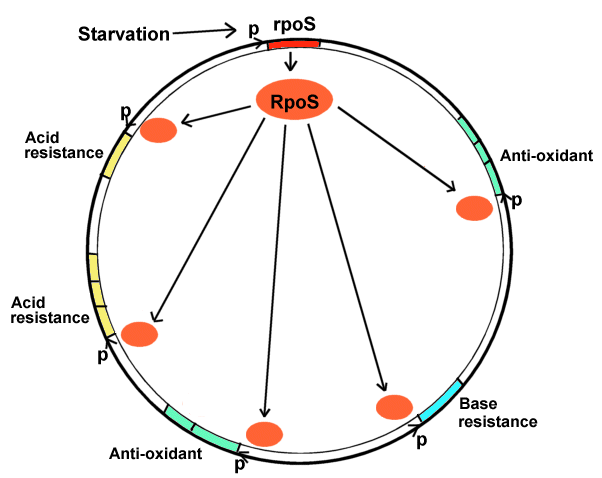 When bacteria enter your digestive tract, how do they adjust to your body's defense mechanisms such as acid and antioxidant stress? When bacteria exit the intestine how do they cope with starvation?
Research on Operons
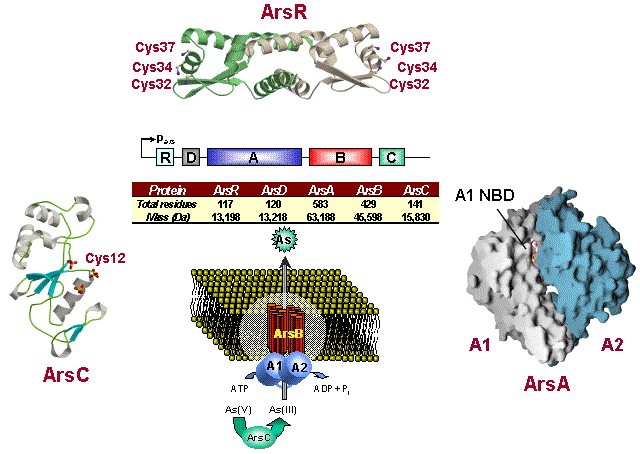
An example of an arsenic resistance operon from Barry Rosen's Arsenic Research Lab.
M. Donnenberg, U. Maryland 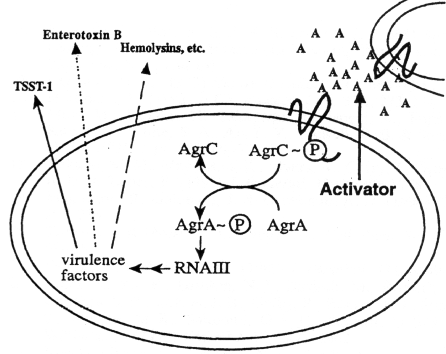
Balaban & Novick, 1995, PNAS 92:1619.
>> Solutions to Problems >> |