Biology Dept Kenyon College |
Eukaryotic Gene Control |

|
Biology Dept Kenyon College |
Eukaryotic Gene Control |

|
| Eukaryotic
RNA polymerases; in vitro transcription
Enhancer Elements on DNA Transcription Factors and Transcriptional Activation Post-transcriptional Control Advanced Topic: Research on the Gridlock Transcription Factor Eukaryotic Gene Control Eukaryotic control sites include promoter consensus sequences similar to those in bacteria.
However, there
can be many
control sequences, called enhancers and silencers, responsive to many
different
signals. Enhancers were defined by cis/trans complementation
experiments,
in which their activation only occurred when they were present on the
same
DNA helix with the gene under their control. Thus they were
originally
called cis-acting elements; this terminology is still used in
experiments
defining new regulatory sites.
Three RNA Polymerases in Eukaryotes Review from before break: Eukaryotes have three different RNA polymerases, which transcribe three different classes of genes. RNA pol II transcribes hnRNA (precursor to mRNA). RNA pol I and III transcribe functional RNAs such as rRNAs and tRNAs. Initiation of RNA pol II transcription requires multiple basal transcription factors. Most of these were identified initially through biochemical approaches, i.e. fractionation of nuclear extracts (by chromatography or density gradient centrifugation) and reconsitution of transcription in vitro. For example, in this experiment, different purified basal transcription factors (TBP, TFIIB, IIF, IIE, IIH) and RNA polymerase II were mixed and matched to see which would support transcription from the adenovirus major late promoter.
(Shilatifard A, Haque D, Conaway RC, Conaway JW. J Biol Chem 1997 Aug 29;272(35):22355-63)
Discovery of Enhancers: Using recombinant DNA transfected into cultured cells. Susumu Tonegawa: Transcription of the human antibody heavy chain gene is under control of enhancer elements in Intron 1.
Assessment
of enhancer elements using recombinant reporter genes in transgenic
mice:
How do enhancer elements work to regulate transcription of specific genes in specific times and places? By serving as binding sites for transcription factors--proteins that regulate transcription.
DNA:Protein and Protein:Protein interactions are important for transcription factor function. Note modular structure of transcription factors: one part of the protein is responsible for DNA binding, another for dimer formation, another for transcriptional activation (i.e. interaction with basal transcription machinery). Dimer formation
adds an extra
element of complexity and versatility. Mixing and matching of
proteins
into different heterodimers and homodimers means that three distinct
complexes
can be formed from two proteins.
A COMPREHENSIVE MODEL OF REGULATION OF RNA POLYMERASE II TRANSCRIPTION: Although they are cis-acting, the enhancers and silencers can be strung out across 10-20 kilobases (thousands of base pairs) of DNA upstream. Some signals can even be downstream of the coding gene, or even found within introns (!) How can this be possible? Long regions of the DNA can loop over to enable the regulatory connections.
Based on Robert Tjian, "Molecular Machines that Control Genes," Scientific American.
RECALL. . . Splicing of hnRNA to make mRNA
Post-transcriptional
control Degradation of
mRNA.
Certain hormones can stimulate (or retard) the rate of degradation of
mRNA,
thereby decreasing (or increasing) its availability fortranslation to
protein.
Post-translational control Protein
cleavage and/or
splicing. The initial polypeptide can be cut into different
functional
pieces, with different patterns of cleavage occurring in different
tissues.
In some cases, different pieces may be spliced together.
Problem. Gene expression is subject to many levels of control. Outline the mechanisms at all the levels of control, from gene sequence to final product. Explain why control is useful at different levels. Research Advanced topic: "Gridlock," a Developmental Regulator in Zebrafish Zebrafish is a
major model
system for vertebrate development.
Genotype and phenotype of grl.
When grl mRNA was injected, the mutant embryo developed normal arterial circulation! 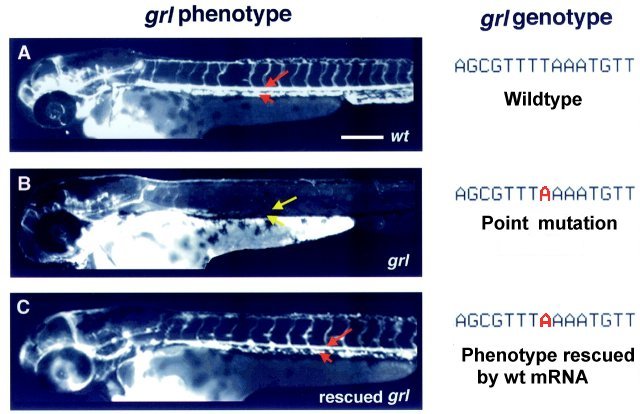 Zhong et al, 2000, Science 287:1820 How was grl
mapped,
located, and sequenced?
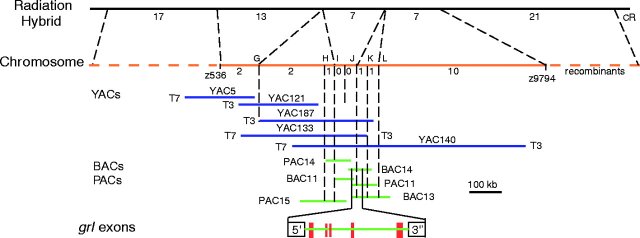
Zhong et al, 2000, Science 287:1820
The terminal ends of the protein form a clamp that binds DNA. Major or minor groove? Check the Chime model! The protein encoded by grl appears to be a transcriptional repressor that distinguishes certain populations of aortic angioblasts (precursor cells of arterial structures). |