1. For a restriction enzyme of sequence CAATTG, about how many cut sites would you expect in the entire (human) genome?
Four
possible bases at each of six positions = 1/46 possible combinations.
The human genome has 3 x 109 base pairs.
The number of CAATTG sites = 3 x 109 bp /46 bp per
site = approx. 730,000
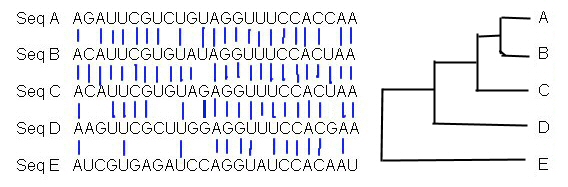
2. Sequences to align, and sketch a likely divergence tree:
3.
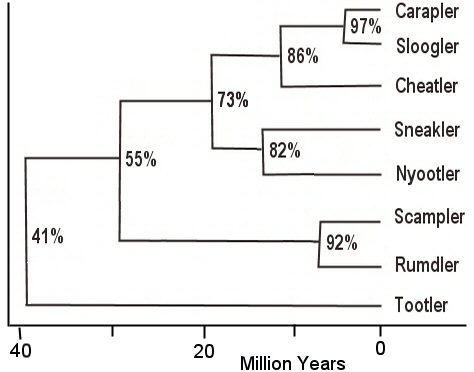
Draw an evolutionary tree of these animals from planet Zlerin. Assume
the species mutate and diverge at 1.5% / million years:
Table shows percent relatedness of each pair of species.
|
|
|
A |
B |
C |
D |
E |
F |
G |
H |
| A |
Carapler |
100 |
86 |
73 |
55 |
55 |
97 |
73 |
41 |
| B |
Cheatler |
86 |
100 |
73 |
55 |
55 |
86 |
73 |
41 |
| C |
Nyootler |
73 |
73 |
100 |
55 |
55 |
73 |
82 |
41 |
| D |
Rumdler |
55 |
55 |
55 |
100 |
92 |
55 |
55 |
41 |
| E |
Scampler |
55 |
55 |
55 |
92 |
100 |
55 |
55 |
41 |
| F |
Sloogler |
97 |
86 |
73 |
55 |
55 |
100 |
73 |
41 |
| G |
Sneakler |
73 |
73 |
82 |
55 |
55 |
73 |
100 |
41 |
| H |
Tootler |
41 |
41 |
41 |
41 |
41 |
41 |
41 |
100 |
4. Explain how the processes of gene cloning and PCR amplification are based on naturally occurring enzymatic processes in microbial genetics.
Gene
cloning involves cutting a plasmid and target DNA with a restriction endonuclease,
a type of enzyme made by bacteria in order to cleave foreign DNA (such
as from a phage) that endangers their survival. The plasmid DNA incorporates
the cleaved target DNA into its sequence by annealing (hybridization)
of its staggered ends, regenerating the restriction site sequences. Completion
of the recombinant plasmid requires use of ligase to seal the phosphodiester
backbone. Ligase is an enzyme used by bacteria to seal the nicks between
portions of the lagging strand of DNA replication.
PCR amplification involves the use of a single enzyme, thermostable DNA
polymerase, which can withstand the heating and cooling cycles of repeated
amplification and denaturation of a small DNA molecule. The DNA polymerase
is obtained from thermophilic bacteria or archaea.
5. Explain the role of nucleic acid hybridization in identifying particular gene sequences within genomic DNA or cellular RNA. Compare and contrast the use of Northern blot, real-time PCR, and microarrays to measure gene expression.
Nucleic acid hybridization involves the annealing (hydrogen-bond pairing of bases) between two strands of DNA or RNA from different source molecules. Genomic DNA can be analyzed by Southern blot to identify a specific sequence on a DNA fragment of defined length, resulting from restriction endonuclease cutting. The target DNA is genomic (or other source) DNA cleaved with a restriction enzyme and separated through an electrophoretic gel. The probe DNA is a gene fragment (radio-labeled or fluorescent-labeled) that will hybridize to its complementary sequence in the genomic DNA.
Cellular mRNA can be analyzed by techniques that identify one specific mRNA expressed from one gene. Northern blot uses a single specific cDNA probe to detect the presence of mRNA encoded by a specific gene. A microarray contains an array of short DNA probes that may hybridize to many specific gene sequences; in some cases all the known genes of a genome. In microarray use, the target is usually not RNA but cDNA. Real-time PCR measures the quantity of a specific DNA or mRNA sequence by the timing of amplification cycles. Real-time PCR involves initial target hybridization to a pair of primer ends, not the whole gene sequence. There follow cycles of denaturation, synthesis, and rehybridization (reannealing). The key principle of real-time PCR is that the more copies of a given DNA in a sample, the earlier the curve of amplification starts to rise. The amount of DNA (usually cDNA, based on mRNA) is thus measured negatively in cycle numbers (number of PCR amplification cycles). See this image.
6. Explain examples of regulating gene expression at the level of DNA sequence; RNA transcription; protein translation; and modification of RNA and of proteins.
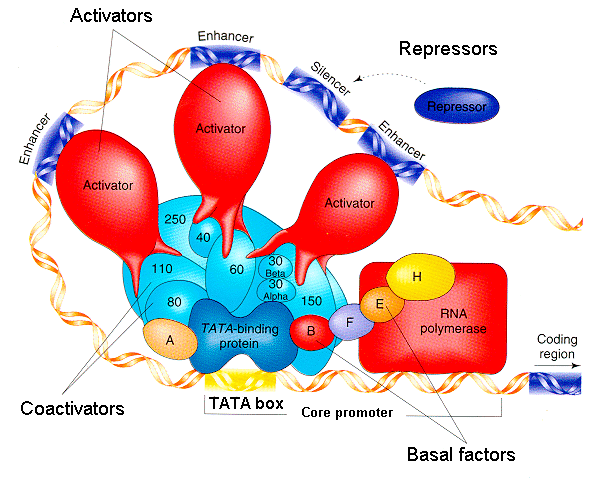
7. Explain with a diagram the comprehensive model of eukaryotic transcription regulation. Explain which features are analogous to specific features of bacterial gene regulation.
Regulation of eukaryotic transcription requires: basal factors, protein complexes vthat enable RNA polymerase to transcribe from the core promoter; coactivators, protein complexes that connect the basal factors to the protein-binding domains of the activators; transcription factors (activators) that contain protein-binding domains to bind the coactivators, and DNA-binding domains to bind the enhancer sequences on DNA. The basal factors are analogous to bacterial sigma factors. The transcription factors are analogous to bacterial activators such as CAP.
Diagram:
8. Explain with examples the typical structure of eukaryotic transcription factors, including DNA binding and signal reception domains.
9. Explain the difference between operon and regulon. What kinds of molecular mechanism typically mediate the control of a regulon?
10. For each partial-diploid for the lac operon, explain whether B-galactosidase is expressed, and whether lactose is needed as inducer, and why:
| A. p+
o+ lacZ+ |
lacI- |
B. p- oc lacZ+ |
lacI- |
A. B-galactosidase cannot be expressed by the first operon because of a mutant structural gene. It is expressed by the second operon, and will require lactose inducer to relieve repression by repressor.
B. B-galactosidase can only be expressed by the operon with a functional promoter (p+). Expression will not require lactose, because neither lacI operon is functional, therefore no repressor is made.