Test 2 BIOL 263, 2007
Name
____________________________________
Each question 20 points.
1. Sketch a labeled diagram of the pathway described in this abstract. Include as many molecular interactions as possible. Explain how the gene expression is regulated.
Abstract
p53 is a transcriptional activator that prevents tumors by activating tumor supressor genes. p53 activity is regulated by numerous post-translational modifications, including lysine methylation. Histone lysine methylation has recently been shown to be reversible; however, it is not known whether non-histone proteins (such as p53) can be demethylated. Here we show that, in human cells, the histone lysine-specific demethylase LJD2 interacts with p53 to repress p53-mediated transcriptional activation and to inhibit the role of p53 in promoting apoptosis. We find that, in vitro, LJD2 removes both monomethylation (K370me1) and dimethylation (K370me2) at K370, a previously identified Smyd2-dependent monomethylation site. However, in vivo, LJD2 shows a strong preference to reverse K370me2, which is performed by a distinct, but unknown, methyltransferase. Our results indicate that K370me2 has a different role in regulating p53 from that of K370me1: K370me1 represses p53 function, whereas K370me2 promotes association with the coactivator 53BP1 (p53-binding protein 1) through tandem Tudor domains in 53BP1. Further, LJD2 represses p53 function through the inhibition of interaction of p53 with 53BP1. These observations show that p53 is dynamically regulated through lysine methylation (by Smyd2) and demethylation; and that the methylation status at a single lysine residue confers distinct regulatory output. Lysine methylation therefore provides similar regulatory complexity for non-histone proteins and for histones.
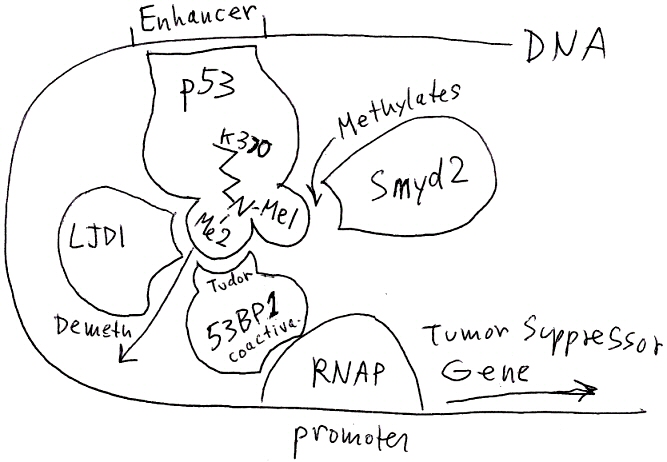
p53 binds an enhancer sequence on the DNA. Its second methyl on lysine K370 promotes binding to coactivator 53BP1 (which could be part of the "mediator complex"). The complex activates transcription of a tumor suppressor gene. Smyd2 methylates K370 (Me1), and an unknown methylase adds the second methyl (Me2). LJD1 demethylates p53-K370-Me2, thus preventing p53 from interacting with 53BP1, and repressing transcription.
2. Find which disease is caused by mutation of the
human MYBPC3 gene. What
is the function of the gene product? How might the gene product interact
with other cell components?
State the human chromosome, the chromosome, and name the nearest known
genes on either side.
Does this gene have a homolog in the mouse chromosome?
If so, find its name in the mouse, chromosome, and the genes on either
side.
MYBPC3 is the cardiac-type Myosin-binding protein
C. A defect causes hypertrophic cardiomyopathy (abnormal growth of heart
muscle). The protein binds myosin heavy chain and is phosphorylated (by
a kinase) to induce muscle contraction. The gene encoding this protein
is found on human chromosome 11, between the MADD gene and the SP11 gene.
In the mouse, the ortholog MYBPC3 is on chromosome 2, in an inverted region
that includes flanking genes Sfpi1 and Madd.
3. Explain the advantage and limitations of each of these
measures of gene expression in microarray data.
Expression
index:
The magnitude of expression of a gene in one biological sample, based
on weighted analysis of the probes for that gene. Expression indices indicate
the amount of gene expression compared to the amount of other genes in
the same sample. They can suggest the likely amount of error (the smaller
the expression index, the greater the noise contribution). In themselves,
expression indices indicate no comparison with other biological samples
or conditions.
Expression
difference:
The expression difference subtracts one expression index of a gene from
one sample from an expression index of the same gene in another sample.
It indicates the quantity of expression difference, but does not indicate
the relative proportions of the two expression levels, which is more likely
to have biological relevance.
Expression
ratio:
The ratio between expression indices for a gene from two biological samples
provides a good measure of possible up- or down-regulation of transcription
between two conditions. It provides no perspective however on the overall
sampling error and the overall level of transcription in the two samples.
Log2(ratio):
The log2 of the expression ratio represents the expression ratio in a
manner such that positive and negative values appear equivalent; that
is, a 1:2 ratio appears equivalent in magnitude to a 2:1 ratio. It still
provides no correction for sample error and for overall transcription
levels.
Centered
log2(ratio)
The centered log2 of the expression ratio provides an expression ratio
normalized to the overall levels of transcription of all genes in two
conditions. This value is commonly presented as the most biologically
relevant. However, it eliminates information about about overall transcription
levels which might be relevant under conditions where a large number of
genes are co-regulated; for example, down-regulation of of the entire
protein synthesis apparatus (typically about 40% of a bacterial cell metabolism).
4. Identify this type of transcription
factor domain.
Explain its molecular structure, including how specific amino acids hold
it together, and how it specifically binds DNA.
This complex contains a helix-loop-helix domain
of a transcription factor bound to DNA. Each peptide of the dimer contains
a long recognition helix contacting the DNA major groove, then a loop,
then another alpha helix that
binds to the partner chain.
The
protein-protein contacts include four pairs of leucines that interact
with Van der Waals forces. The recognition helix-DNA contacts include
lysines and arginines whose positive charges interact with phosphates,
and a histidine that interacts with the base pairs of the major groove.
5. Explain how one specific antibiotic binding to specific parts of the ribosome provides information about the A-to-P site transition.
Explain why this antibiotic only affects prokaryotes, not eukaryotes, despite the high conservation of the ribosome throughout living organisms.
Spectinomycin is known to inhibit translocation of peptidyl-tRNA from the A-site to the P-site. The position of spectinomycin within the ribosome was visualized by difference Fourrier analysis comparing the 30S subunit crystallized with and without the antibiotic. The position of spectinomycin was located at 16S helix 34, near helix 35 (according to Carter et al, 2000). This position is not at the A- or P-site, but is lies near the proposed "pivot point" of the ribosome "head." The antibiotic is proposed to inhibit translocation by preventing the "head" from shifting position. This finding confirms the view that the ribosome functions as a whole, with conformational change occurring throughout the complex, not just at the binding sites for its RNA and amino-acyl-tRNA substrates.
[Other answers possible for streptomycin or paromomycin.]
Despite the overall conservation of ribosome structure and function, there are subtle differences between the prokaryotic and eukaryotic structures. These subtle differences can determine the precise size of potential binding sites for antibiotics. As a result, most antibiotics affect either the prokaryotic ribosome or the eukaryotic ribosome, not both.
Grade range:
94-96% A+
90-93% A
85-89% A-
80-84% B+
76-79% B
71-75% B-
65-70% C+