BIOL 263 Test 1 2007 Total points: 100 Do not refer to CAP protein, aside from questions on the Benoff article.
1. (10 pts) Explain two different kinds of function that Z-DNA can serve in a living cell. Cite a specific example of one of these functions.
Z-DNA formation converts right-handed B-DNA into a left-handed twist; this occurs in the wake of RNA polymerase unwinding the helix and transcribing to RNA. The formation of left-handed twist temporarily relieves torsional stress.
Z-DNA formation thus can act as a signal of the proximity of nascent mRNA to RNA-editing enzymes such as ADAR1.
Segments of DNA sequence favoring Z-DNA formation (CG rich) may participate in promoters or enhancers for transcription of specific genes.
2. (10 pts) A DNA plasmid of length 10,500 base pairs has a linking number of 950. Assuming one DNA twist covers 10.5 base pairs, how many superturns does the plasmid have? Negative or positive? Show your calculation.
Lk = Tw + Wr Tw = 10,500 bp / 10.5 bp/turn = 1000 turns Lk = 950 = 1000 + Wr Wr = -50 = 50 negative superturns
Explain what class of enzymes could change the number of superturns, and what such an enzyme need to do to DNA.
3. (15 pts) Explain the data shown for a bacterial DNA consensus sequence for a class of sites regulating transcription. Explain: -- What kind of regulator this probably is (activator or repressor) and how it probably interacts with RNA polymerase.
The binding protein is probably an activator which binds a segment in the upstream region (such as UP) and also binds to the C-terminal domain of alpha subunits of RNA polymerase.
-- Why the various sites show the observed trend in transcription levels (approximately).
The efficiency of transcription increases as the promoter sequence approaches that of the consensus sequence.
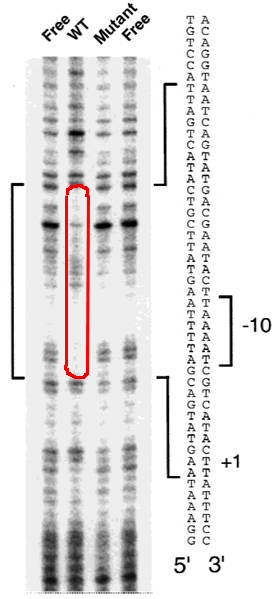
This figure shows the results of a DNA footprint. The DNA samples (free; with protein; with mutant protein) are cleaved with a DNase enzyme that attacks duplex DNA. All DNA is end-labeled so that only segments with the same end will be labeled in the gel. The enzyme cut sites are uneven, but do show distribution across the sequence. The presence of bound protein prevents some of the cut sites, and so certain segment lengths fail to show up in the gel.
What specific conclusions may be drawn by comparison of the “free” DNA, the DNA with a wild-type protein, and the DNA with a mutant (nonfunctional) protein? What is the likely function of the protein?
The wild-type protein binds a region from approximately -10 to -25 bp upstream of the transcription start. The mutant protein fails to bind this region, as its banding pattern looks essentiall the same as that of the free DNA.
The binding protein is probably a repressor, which binds a segment overlapping the core promoter (-10 to -35).
5. (10 pts) Explain two different kinds of bacterial transcriptional regulation that involve RNA secondary structure and do not require protein transcription factors. Give a specific example of each.
Transcription of an operon encoding biosynthesis of an amino acid (such as Trp or His) can be attenuated as a result of high concentration of the amino acyl-tRNA available to the ribosome. When the ribosome readily incorporates the amino acid, it moves swiftly through an mRNA hairpin, allowing formation of a different hairpin close behind the RNA polymerase. The hairpin near the RNA polymerase destabilizes transcription and the RNAP complex falls apart. When the amino acid is scarce, however, the ribosome pauses "waiting" for it, just long enough for a different hairpin to form, which precludes formation of the attenuation hairpin.
A riboswitch consists of a complex RNA pseudoknot that forms only in the presence of a specific biosynthetic product, such as vitamin B12 or thiamine. Riboswitch formation can terminate transcription of the biosynthetic operon.
6. (10) Explain how RNA-RNA interactions contribute to splicing mRNA in eukaryotes.
The splicing reaction
is mediated by the spliceosome complex, which is composed of five or
six subsidiary complexes, each containing an RNA (U1, U2, U3, U4, U5,
U6). The RNA of a sub-complex can hybridize to the upstream or downstream
end of the intron, marking it for splicing (U6 and U2). The two sub-complex
RNAs then hybridize to each other, to bring together the two ends of
the intron for the lariat reactions:
How is splicing regulated?
Splicing is regulated by splicing activators and repressors.
What biological function is served by splicing?
Splicing allows combination
of different variants of different exons, to form slightly different
versions of a protein. Splicing provides an enormous amount of increased
diversity for the diverse needs of different tissues and developmental
stages of a multicellular animal or plant. Questions based on Benoff et al (2002) 7. (10 pts) How were the CAP components crystallized, and how was X-ray
data obtained? 8. (10 pts) What are the molecular components of the unit cell, and how many of each? Describe specific DNA sequences relevant to complex formation. 9. (10 pts) Identify two specific interactions between the alpha protein carboxy-terminus and DNA. Explain how each specific interaction is a “weak force” mediates the interaction.
|