So far we have discussed reassortment and
recombination
of alleles.
Now we discuss “allele conversion” by
mutation.
Mutation is
change in DNA sequence that is inherited by offspring.
A mutation event
is
how the allele sequence changes. Two things must happen:
-
A change in the molecular structure of DNA
-
Failure of editing enzymes to correct the
change; copying
into new DNA
Mutant strain: A
population of descendents of the individual in which the original
mutation
event occurred. The “mutation” is now inherited by
the regular reassortment
and recombination mechansisms, as are other alleles.
Rate of mutation:
How
often a given map position mutates. In practice, this is hard to
measure.
Frequency of mutation:
What percent of alleles contain a
sequence defined
to be “mutant,” in a given population at a given point in
time. This
is easy to measure.
Note: In nature there is no such thing as
“wild type.”
All existing alleles are the result of the past 4 billion years of
mutation
events.
Mutation events are
rare.
How to detect them?
-
Observation of
large numbers of progeny. (Patience.)
-
Positive selection.
For traits which confer survival advantage: Subject them to the
selective
environment. Example: plate bacteria on agar containing an
antibiotic.
-
Negative selection.
For traits which prevent survival,
under a given
condition. Example: Replica plate bacteria colonies on agar
lacking
a nutrient which the “wild type” strain can make with its
own enzymes.
| Problem (1):
Explain how and
why each of the above approaches reveals mutant strains. Compare
the advantages and disadvantages of each. |
Classifying mutations
-
Phenotype.
Appearance;
behavior; auxotrophy; drug resistance; conditional on environmental
factors,
etc.
-
DNA structure.
Gene
mutations vs. Chromosomal mutations. Point mutations: transitions,
transversions
-
Deletion; Insertion;
Inversion
-
Information
effect.
Silent; nonsense; missense; frameshift
Problems--"Spontaneous"
mutations:
(2)
Chromosome mutations: How
do they occur? Review the process of Recombination.
Think of mistakes that can happen, especially with the Holliday
structure,
and with supposedly homologous base pairing.
(3)
Point mutations: How do they occur?
Review DNA replication.
Where can "errors" creep in? |
Note that many different kinds of mutation can
prevent
mRNA transcription, resulting in the same phenotype.
Successive mutations play a major role in the
appearance
and progression of malignant tumors. From the Cornell
University Medical College:
A major factor in progression
appears to
be that most tumor cells are genetically less stable than normal cells
and this instability produces variant clones. Chromosomal abnormalities
in number and structure are often seen in tumor cells. These
abnormalities
include: a gain or loss of chromosomes (aneuploidy);
deletion
(loss of a segment of a chromosome);
inversion
("flip-flop" of two segments of a chromosome); translocation
(rearrangement of segments between two chromosomes); and
mutation (heritable
change in the structure or expression of a gene) ranging from
chromosomal
change to single base-pair substitution
(point mutation). Molecular genetic mechanisms
implicated in
tumor progression include: chromosomal
rearrangements
or mutations that "activate" cell oncogenes
(proto-oncogenes); and
loss of putative "tumor suppressor" genes.
Note: The phenotypic result of a mutation is hard
to predict;
it depends on the physiology of the particular case. The
magnitude
of the mutation may have no correlation with the magnitude of the
phenotype.
For example:
-
A major chromosome inversion may result in
completely normal
phenotype, so long as no genes are lost.
-
A single base pair replacement (point
mutation) may decrease
or eliminate function of a gene, resulting in lethality. Example,
Sickle-cell anemia.
| Problem (4):
Transcribe the
following DNA sequence into RNA:
5' A A T G G G
C T A C T T A
G C C A C T A G G C T T T A G C C 3'
3' T T A C C C G A
T G A A T
C G G T G A T C C G A A A T C G G 5'
You should find
two ways to do
it.
Which way could be
mRNA to be
translated into a short protein? Why?
Write the protein,
using the
genetic code in your text.
(Note however:
Real coding sequences
would have hundreds of base pairs.)
Why cannot the
other RNA encode
an entire protein?
Perform the
following types of
"mutation" in your DNA sequence:
Frame shift
Base pair
substitution
Silent mutation
Missense mutation
Nonsense mutation
Show the
protein that would result
from each mutation.
|
Mutagenesis
Certain chemicals and environmental factors may
increase
the rate of mutation. These are called mutagens.
The Ames test provides a rough measure of the effect of a
chemical
mutagen.
Mutagens include:
-
Base analogs which
incorporate into DNA and pair incorrectly. An example is 5-bromouracil.
-
Chemicals which modify
existing
bases of DNA and cause incorrect pairing.
-
Intercalating agents
are
base pair analogs which intercalate
between
bases, resulting in addition
or
loss of a base pair. Lead to frame shift
mutations. An example is the acridine derivative
proflavin:
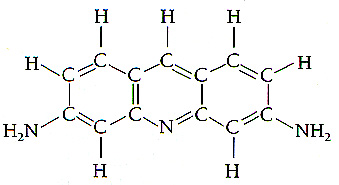
-
High-energy electromagnetic radiation (UV
or gamma rays) cause breakage of the backbone, or
cross-linkage
of bases, such as thymine dimers,
TT
or TC.
Biological mutagenesis is caused by:
-
Mutator genes.
Mutations
within genes responsible for editing lead to high frequency of
mutations
throughout the genome.
Transposition.
Transposable elements can insert
into a chromosome,
excise themselves out of the chromosome, or copy themselves into new
locations.
Transposition is mediated by enzymes, such as a transposase.
Solutions
to problems
Finding
Mutants
| Positive
selection for Gain-of-function
mutations:
Spread organisms on petri plate of media containing antibiotic.
Only
those with antibiotic-resistance mutation will produce colonies.
Negative
selection for Loss-of-functionmutations:
Grow
large number
of colonies. Replica plate or pick colonies onto non-permissive
growth
medium. Whichever colonies fail to grow, go back to original
source
to obtain mutant strain.
|
Are gain-of-function
alleles
dominant or recessive? What about loss-of-function?
Case
Example:
Stickler's Disease
|
Phenotype class
|
Dominant |
Recessive |
|
Gain-of-function
|
New function covers up "normal" version.
Examples:
Huntington's
disease
Drug resistance pump
|
Haploinsufficiency:
One copy of the new
function is
not enough.
Example:
Sickle
cell anemia
|
|
Loss-of-function
|
Haploinsufficiency
ofwild-type
allele.
Usually co-dominant.
Examples:
Stickler's
disease.
Homeotic fly gene,
makes leg instead
of antenna.
|
Loss of function is
covered up by wild-type
allele producing enough functional protein.
Example:
Cystic
fibrosis
|
Find your own disease in OMIM.
Solutions to problems
|


