|
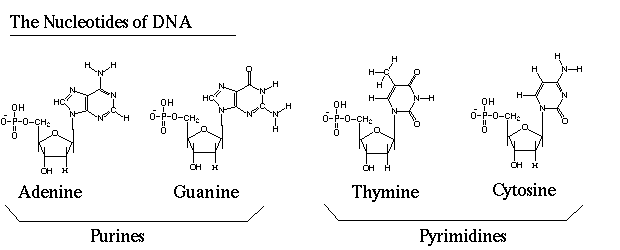
BIOL 114 Test 1 Name ________________________________ Total possible: 50 1. (6 pts) Sketch the structure of one deoxyribose-phosphate unit of a DNA backbone. Indicate the link to the nitrogenous base.
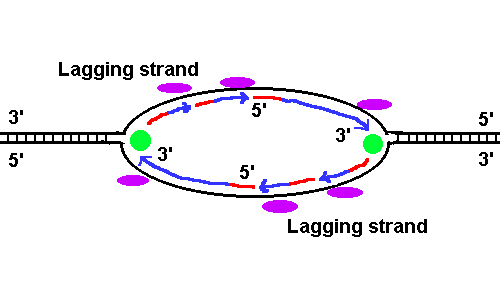
3. (10 pts) Outline all the steps of DNA replication in detail. Include one diagram showing two replicating forks. See DNA Replication.
1. Helicase enzyme unwinds DNA. This reaction needs ATP. At each replicating fork, the exposed single-stranded DNA is protected by single-strand binding proteins (ssb). Primase enzyme binds, preparing to make RNA primers. 2. Primase enzyme makes RNA primer molecules. Each primer hybridizes (base pairs) with DNA, at the origin of replication. The 3' OH end will attach new deoxy nucleotides (dNTPs). The primers will each start a leading strand, 3. DNA polymerase III (Pol III) attaches new dNTPs to the 3' OH end of the growing chain of the leading strand, which elongates toward the replicating fork, 5' to 3'. (For each origin, there are TWO leading strands; why?) For each NTP, a pyrophosphate (PP) is released, providing the necessary energy. 4. More primers hybridize to the opposite strand of DNA. Pol III starts elongating 5' to 3' but it keeps running into the back of an RNA primer. This is the lagging strand. There are TWO lagging strands (why?) 5. DNA polymerase I (Pol I) starts at “nicks” in the growing strands. It edits the strand by removing bases ahead of it (5' end), including RNA and mismatched bases, while elongating the strand "behind" 5' to 3'. It replaces all RNA nucleotides with dNTPs. 6. Ligase seals the phosphate bonds at all “nicks” in the DNA. 7. Editing endonucleases excise mismatched nucleotides, replacing with the proper match. How do they know which is old DNA vs. new DNA? The old DNA contains methyl groups on some of its cytosine bases. 8. Gyrase restores negative superturns in DNA. ATP is needed. 9. Methylases add methyl groups to the new DNA, at the same positions as the original strands. Now the two daughter helices are indistinguishable from each other, and from the original helix. 4. (6 pts) Explain what
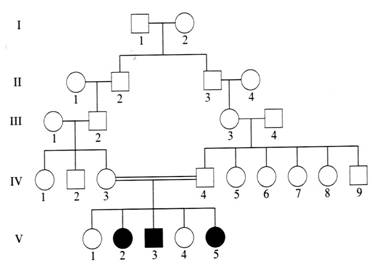
is Z-DNA, and what is its main role in living systems. 5. (12 pts) For each observation, explain the genetic basis and propose a way to test: A. Fruit flies are born with legs instead of antennae (antennapedia trait). No pure-breeding strain can be obtained, however; some wild-type offspring always occur. The double allele of antennapedia is lethal; single allele is codominant with wild-type. To test, cross wild-type with antennapedia. Result of cross: 1/2 wild type, 1/2 antennapedia. When two antennapedias are crossed, ratio will always be 2/3 antennapedia, 1/3 wild type (the 1/4 "pure" antennapedia dies before birth.) B. Two bay (brown) horses are intercrossed, yielding mostly bay (about 9/16) but also a few black and sorrel horses. An occasional mouse-colored horse appears. The two bays carry two different color loci. When both colors are present, they combine to appear bay (brown.) When neither is present, the color appears pale (mouse-colored). Two dihybrid bays crossed produce a 9:3:3:1 ratio. Black crossed with sorrel should produce bay. A dihybrid bay crossed with mouse-colored (test cross) should produce all four colors, 1:1:1:1. 6. (4 pts) State the most probable basis for inheritance of the trait shown in this pedigree, with a brief explanation. Autosomal recessive inheritance is the most likely result of this consanguinous marriage. Both males and females inherit the trait from parents who do not show it, but both parents carry it from a common ancestor in generation I.
7. (8 pts) A corn breeder investigating disease resistance (R = resistant; r = sensitive) and tassel length (T= long; t = short) test-crosses a corn plant that is dihybrid for disease resistance (R/r) and long tassels (T/t). The resulting corn plants are: 9 resistant; long tassels 7 sensitive; short tassels 90 resistant; short tassels 94 sensitive; long tassels Explain these results, with appropriate calculations. Explain the genotypes of the parents of the original dihybrid crossed above (i.e. the grandparents of the offspring shown.) Recombinant percentage = (9+7)/(9+7+90+94) = 0.08 = 8% = 8 map units distance between the genes.The parental phenotype proportions indicate that the grandparent genotypes were (1) Rt/Rt, (2) rT/rT. |