Biology Dept Kenyon College |
|

|
Biology Dept Kenyon College |
|

|
| Molecular
Evolution
Evolution of Color Vision Genome and Proteome Molecular Evolution The study of molecular evolution--how gene sequences change and evolve over time--is an important part of bioinformatics, the study of DNA sequence information.
Spontaneous
mutations occur in different individuals.
Different
individuals pass on different mutations to their offspring.
The
mutations most likely to remain without negative selection are silent
or neutral mutations.
In current populations, we cannot distinguish between the original sequence and the versions that mutated. However, by comparing DNA from current populations, we can calculate the most probable sequence of the original. Examples:
Molecular evolution can be used to measure divergence of species. For example: 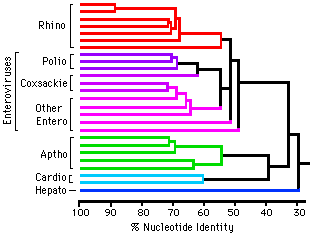 Picornoviridae This dendrogram shows the relative time of divergence of different species of virus, including Apthovirus, the cause of foot and mouth disease (in the news--devastating cattle in England and Europe.) Genes can diverge: Example: Homeobox genes were originally discovered in Drosophila (more later in Development section of BIOL 14).
The
original homeobox
(Hox) gene in invertebrates duplicated and evolved into several
paralogs.
Homeobox
Genes--A set of transgenic mice containing Hox gene "knockouts"
A homeobox gene HOX-A1 has been implicated as a key to autism. See Patricia Rodier's article on Autism in Scientific American (February 2000).
Molecular Biology of Color Vision -- HHMI Tutorial
Dr. Jeremy Nathans cloned a bovine rhodopsin gene obtained from cDNA from cow retinas. He then used the bovine gene -- an ortholog of human rhodopsin -- as a probe to isolate clones of the red, green, and blue opsin genes (paralogs), using his own genomic DNA. (What kind of hybridization blot?) He had to test his own color vision first, to make sure he was not color-deficient (estimates range from 7-30% of the male population.) The color
receptor
opsins all arose by gene duplication
and subsequent divergent evolution
into paralogs of different function. Because the green opsin gene
has a duplicate copy tandem to the red opsin gene, on the X chromosome,
mistaken crossover can occur between these two genes. The result
is failure to distinguish red from green; or in rare cases, perception
of an anomalous color. In extremely rare cases, a female carrier
of such a gene has been reported to "see" a fourth color.
Genome and Proteome One of the most exciting applications of gene cloning, PCR, and rapid sequence analysis is the Human Genome Project. Several other animal, plant, and microbial genomes have now been sequenced. The human genome will take a bit longer, but we already have expressed sequence tags for most of the protein-encoding genes. Human
mutations in
disease: See
Online Mendelian
Inheritance in Man
We can
learn a lot
about human genes by studying orthologs from model systems such as
The
Model Worm -- C. elegans.
There are numerous ethical questions to consider; for example, whose DNA will be sequenced? What ethnic classes will be considered "standard" or "normal"? Will we neglect alleles of significance to certain ethnic groups? Once a genome is sequenced, we still have to figure out how it gets expressed, and what all the proteins and RNAs are doing in the cell. The proteome, the collection of all proteins made by a cell, is a far more daunting challenge than the genome.One tool to study the proteome is 2D gel electrophoresis. This is a two-step procedure:
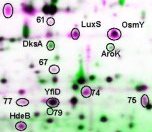
For human examples, see the database at NIMH-NCI. |